|
Size: 779
Comment:
|
Size: 8945
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial: Sources estimation = This tutorial is still based on Sabine Meunier's somatotopy experiment, called ''TutorialCTF ''in your Brainstorm database. |
= Tutorial 7: Source estimation = Now you have in your database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the recordings. Many solutions are described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust. |
| Line 4: | Line 4: |
| Now you have in you database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the results. Many solutions described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust. | <<TableOfContents(2)>> |
| Line 6: | Line 6: |
| For more information about inverse models and minimum-norm estimation, visit the [[Theory]] page. | == Computing sources for a single data file == 1. Right-click on ''Subject01 / Right / ERP'' > ''Compute sources''.<<BR>><<BR>> {{attachment:popupComputeSources.gif}} --- {{attachment:panelComputeSources.gif}} 1. With this first window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation. The default "Normal mode" only let you edit the following options:<<BR>> * '''Comment''': This field contains what is going to be displayed in the database explorer. * '''Method''': Please select wMNE. The other methods dSPM and sLORETA are aloso based on wMNE. They may give better and/or smoother results depending on the cases. * '''Sensors type''': Modalities that are used for the reconstruction. Here we only have one type of MEG sensors (axial gradiometers), so nothing to change. * '''Expert mode''': Show many other options we really do not care about right now. This will be explained in another tutorial. * Click on Run. 1. A new file is available in the database explorer.<<BR>><<BR>> {{attachment:treeMinNorm.gif}} * It is displayed'' inside ''the recordings file ERP, because it is related to this file only. * Meaning of that weird filename: "MN" stands for "Minimum Norm", and "Constr" stands for "Constrained orientation" of the dipoles (the estimated dipoles are considered as normal to the cortex). * You can have a look to what is there in the corresponding matrix file (right-click > File > View file contents). You would find all the options of forward and inverse modelling, and only one interesting field : '''!ImagingKernel''', which contains the inversion kernel. It is a [nVertices x nChannels] matrix that has to be multiplied with the recordings matrix in order to get the activity for each source at all the time samples.<<BR>><<BR>> {{attachment:resultsMat.gif}} 1. Do the same for the ''Left / ERP'' file == Sources visualization == There are two main ways to display the sources: on the cortex surface and on the MRI slices. === Sources on cortex surface === 1. Double-click on recordings ''Right / ERP'', to display the time series (always indispensable to have a time reference). 1. Double-click on sources ''Right / ERP / MN: MEG''. <<BR>>Equivalent to right-click > Cortical activations > Display on cortex. 1. Go to the main peak around 45ms (by clicking on the times series figure)<<BR>><<BR>> {{attachment:sources1.gif}} 1. Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recordings figures: rotation, zoom, ''Surface ''tab(smoothing, sulci, resection...), colormap, sensors, pre-defined orientations (keys from 0 to 7)... 1. Only three new controls are available in the ''Surfaces ''tab, in panel ''Data options'': * __Amplitude__: Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed. * __Min. size__: Hide all the small activated regions, ie. the connected color patches of the less that "min.size" vertices. * __Transparency__: Change the transparency of the sources on the cortex. 1. Take a few minutes to understand what this threshold value represents.<<BR>> * The colorbar maximum depends on the way you configured your ''Sources ''colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 60 pA.m. * On the screenshot above, the threshold value was set to 34%. It means that only the sources that had a value over 0.34*55 = 18.7 pA.m were visible. * If you set the threshold to 0%, you display all the sources values on the cortex surface; and as most of the sources have values close to 0, the brain is mainly blue. * Move the slider and look for a threshold value that would give you a really focal source.The following figures represent the sources activations at t=46ms respectively with threshold at 0% and 90%.<<BR>><<BR>> {{attachment:threshold0.gif}} {{attachment:threshold90.gif}} * The figure on the right shows the most active area of the cortex 46ms after an electric stimulation of the right thumb. As expected, it is localized in the left hemisphere, in the middle of post central gyrus (projection of the right hand in the primary somatosensory cortex). === Sources on MRI (3D) === 1. Close all the figures (''Close all'' button). Open the time series view for Right / ERP. 1. Right-click on Right / ERP / MN: MEG > Cortical activations > Display on MRI (3D). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.<<BR>><<BR>> {{attachment:sources3D.gif}} 1. The sources values are smoothed after being re-interpolated in the volume. The size of the smoothing kernel is configurable: right-click on the figures > MRI display > Smooth... 1. This view can be used to generate contact sheets in a given orientation. Right-click on the figure > Snapshot > Contact sheet (axial): <<BR>><<BR>> {{attachment:contactAxial.gif}} === Sources on MRI (MRI Viewer) === 1. Right-click on Right / ERP / MN: MEG > Cortical activations > Display on MRI (MRI Viewer). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to move the slices (sliders, mouse wheel, click on the views), move in time, change the threshold.<<BR>><<BR>> {{attachment:sourcesMriViwer.gif}} == Computing sources for multiple data files == The sources file we are observing was computed as an ''inversion kernel''. It means that we can apply it to any similar recordings file (same subject, same run, same positions of sensors). But in our TutorialCTF database, the ''MN: MEG'' node only appears in the the ''ERP ''file, not in the ''Std ''one. What is nessary to share an inversion kernel between different recordings ? 1. Compute another source estimation: but instead of clicking on the ''Compute sources'' fron the ''ERP ''recordings popup menu (which would mean that you only want sources for this particular recordings file), get this menu from the ''Right'' condition. This means that you want the inversion model to be applied to all the data in the condition.<<BR>><<BR>> {{attachment:popupComputeMulti.gif}} 1. Select "Minimum Norm Imaging", click on Run. 1. Three new nodes are available in the tree:<<BR>><<BR>> {{attachment:treeMinNormMulti.gif}} 1. The actual inversion kernel you have just computed (1), which contains the same information as the one from the previous section (Computing sources for a single data file). Note that you cannot do anything with this file: if you right-click on it, you will see that there are no ''Display ''menu for it. * Two links (2 and 3) that allow you apply this inversion kernel to the data files available in this condition (''ERP ''and ''Std''). In their popup menus, there are all the display options introduced in the previous section. * Those links are not saved as files but as specific strings in the database: "link|kernel_file|data_file". This means that to represent them, one should load the shared kernel, load the recordings, and multiply them. * The sources for the ''Std ''file do not have any meaning (it contains information about the averaging); but it illustrates the way a kernel is shared 1. Double-click on both sources files available for ''Right / ERP'', and check at many different times that the cortical maps are exactly the same in both cases. 1. You can estimate the sources for many subjects or conditions at once, as it was explained for the head models in previous tutorial: the ''Compute sources'' menu is available on all the subjects and conditions popup menus. 1. Delete the shared kernel (1), we don't need redundant and confusing information for the next steps. Observe that both links disappear at the same time. == Next == You can now observe the cortical maps estimated from a MEG evoked potential. This information is interesting in itself, but it is an image; it is difficult to compare it rigorously with other subjects or stimulations, or to test statistically some hypothesis. The next tutorial describes how to create and study a [[Tutorials/TutScouts|region of interest]]. |
Tutorial 7: Source estimation
Now you have in your database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the recordings. Many solutions are described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust.
Contents
Computing sources for a single data file
Right-click on Subject01 / Right / ERP > Compute sources.
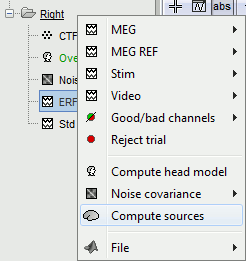 ---
--- 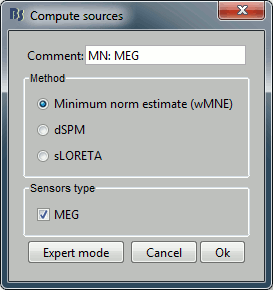
With this first window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation. The default "Normal mode" only let you edit the following options:
Comment: This field contains what is going to be displayed in the database explorer.
Method: Please select wMNE. The other methods dSPM and sLORETA are aloso based on wMNE. They may give better and/or smoother results depending on the cases.
Sensors type: Modalities that are used for the reconstruction. Here we only have one type of MEG sensors (axial gradiometers), so nothing to change.
Expert mode: Show many other options we really do not care about right now. This will be explained in another tutorial.
- Click on Run.
A new file is available in the database explorer.
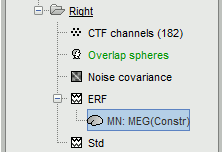
It is displayed inside the recordings file ERP, because it is related to this file only.
- Meaning of that weird filename: "MN" stands for "Minimum Norm", and "Constr" stands for "Constrained orientation" of the dipoles (the estimated dipoles are considered as normal to the cortex).
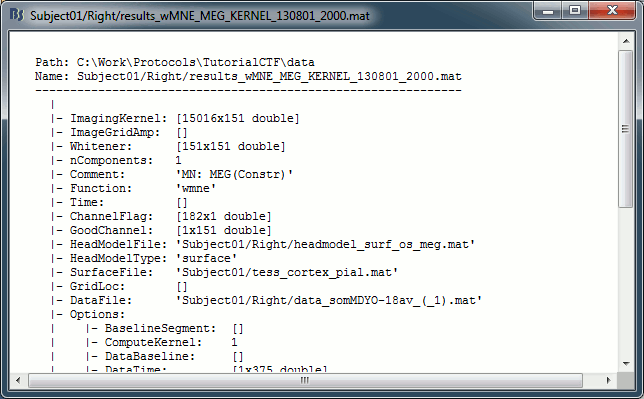
You can have a look to what is there in the corresponding matrix file (right-click > File > View file contents). You would find all the options of forward and inverse modelling, and only one interesting field : ImagingKernel, which contains the inversion kernel. It is a [nVertices x nChannels] matrix that has to be multiplied with the recordings matrix in order to get the activity for each source at all the time samples.
Do the same for the Left / ERP file
Sources visualization
There are two main ways to display the sources: on the cortex surface and on the MRI slices.
Sources on cortex surface
Double-click on recordings Right / ERP, to display the time series (always indispensable to have a time reference).
Double-click on sources Right / ERP / MN: MEG.
Equivalent to right-click > Cortical activations > Display on cortex.Go to the main peak around 45ms (by clicking on the times series figure)
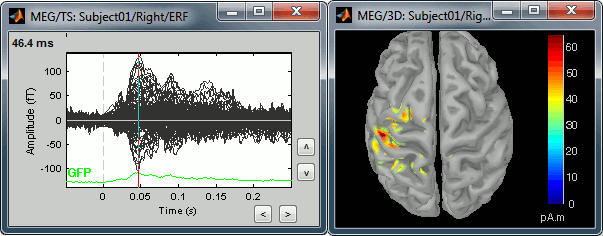
Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recordings figures: rotation, zoom, Surface tab(smoothing, sulci, resection...), colormap, sensors, pre-defined orientations (keys from 0 to 7)...
Only three new controls are available in the Surfaces tab, in panel Data options:
Amplitude: Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed.
Min. size: Hide all the small activated regions, ie. the connected color patches of the less that "min.size" vertices.
Transparency: Change the transparency of the sources on the cortex.
Take a few minutes to understand what this threshold value represents.
The colorbar maximum depends on the way you configured your Sources colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 60 pA.m.
- On the screenshot above, the threshold value was set to 34%. It means that only the sources that had a value over 0.34*55 = 18.7 pA.m were visible.
- If you set the threshold to 0%, you display all the sources values on the cortex surface; and as most of the sources have values close to 0, the brain is mainly blue.
Move the slider and look for a threshold value that would give you a really focal source.The following figures represent the sources activations at t=46ms respectively with threshold at 0% and 90%.
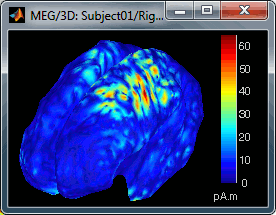
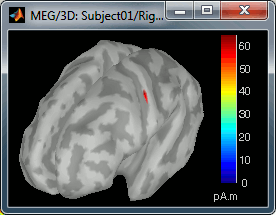
- The figure on the right shows the most active area of the cortex 46ms after an electric stimulation of the right thumb. As expected, it is localized in the left hemisphere, in the middle of post central gyrus (projection of the right hand in the primary somatosensory cortex).
Sources on MRI (3D)
Close all the figures (Close all button). Open the time series view for Right / ERP.
Right-click on Right / ERP / MN: MEG > Cortical activations > Display on MRI (3D).
This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.
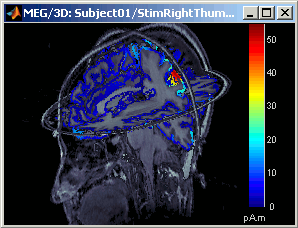
The sources values are smoothed after being re-interpolated in the volume. The size of the smoothing kernel is configurable: right-click on the figures > MRI display > Smooth...
This view can be used to generate contact sheets in a given orientation. Right-click on the figure > Snapshot > Contact sheet (axial):
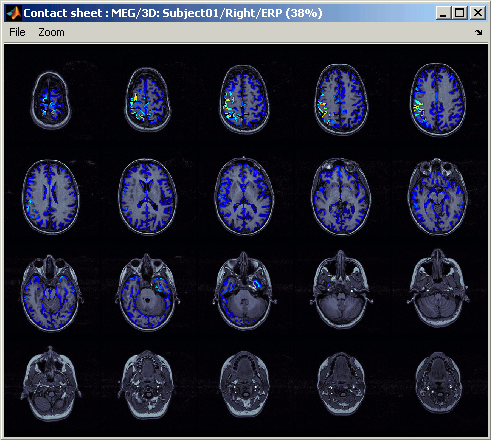
Sources on MRI (MRI Viewer)
Right-click on Right / ERP / MN: MEG > Cortical activations > Display on MRI (MRI Viewer).
This view was also introduced in the tutorial about MRI and surface visualization. Try to move the slices (sliders, mouse wheel, click on the views), move in time, change the threshold.
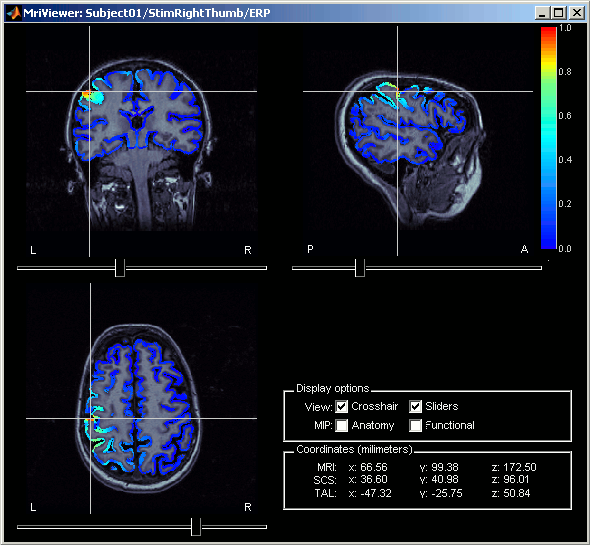
Computing sources for multiple data files
The sources file we are observing was computed as an inversion kernel. It means that we can apply it to any similar recordings file (same subject, same run, same positions of sensors). But in our TutorialCTF database, the MN: MEG node only appears in the the ERP file, not in the Std one. What is nessary to share an inversion kernel between different recordings ?
Compute another source estimation: but instead of clicking on the Compute sources fron the ERP recordings popup menu (which would mean that you only want sources for this particular recordings file), get this menu from the Right condition. This means that you want the inversion model to be applied to all the data in the condition.
- Select "Minimum Norm Imaging", click on Run.
Three new nodes are available in the tree:

The actual inversion kernel you have just computed (1), which contains the same information as the one from the previous section (Computing sources for a single data file). Note that you cannot do anything with this file: if you right-click on it, you will see that there are no Display menu for it.
Two links (2 and 3) that allow you apply this inversion kernel to the data files available in this condition (ERP and Std). In their popup menus, there are all the display options introduced in the previous section.
- Those links are not saved as files but as specific strings in the database: "link|kernel_file|data_file". This means that to represent them, one should load the shared kernel, load the recordings, and multiply them.
The sources for the Std file do not have any meaning (it contains information about the averaging); but it illustrates the way a kernel is shared
Double-click on both sources files available for Right / ERP, and check at many different times that the cortical maps are exactly the same in both cases.
You can estimate the sources for many subjects or conditions at once, as it was explained for the head models in previous tutorial: the Compute sources menu is available on all the subjects and conditions popup menus.
- Delete the shared kernel (1), we don't need redundant and confusing information for the next steps. Observe that both links disappear at the same time.
Next
You can now observe the cortical maps estimated from a MEG evoked potential. This information is interesting in itself, but it is an image; it is difficult to compare it rigorously with other subjects or stimulations, or to test statistically some hypothesis.
The next tutorial describes how to create and study a ?region of interest.
