|
Size: 12297
Comment:
|
Size: 22705
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 6: Source estimation = This tutorial is still based on Sabine Meunier's somatotopy experiment, called ''TutorialCTF ''in your Brainstorm database. |
'''[WARNING: This page is part of the old tutorials, please refer to the [[Tutorials|new documentation]]]'''<<BR>><<BR>><<BR>> |
| Line 4: | Line 3: |
| Now you have in you database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the results. Many solutions described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust. | = Tutorial 8: Source estimation = ''Authors: Francois Tadel, John C Mosher, Richard Leahy, Sylvain Baillet'' |
| Line 6: | Line 6: |
| For more information about inverse models and minimum-norm estimation, visit the RefManual section (under construction). | Now you have in your database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the recordings. Many solutions are described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust. |
| Line 8: | Line 8: |
| <<TableOfContents(2)>> | <<TableOfContents(2,2)>> |
| Line 11: | Line 11: |
| 1. Right-click on ''Subject01 / !StimRightThumb / ERP'' > ''Compute sources''.<<BR>><<BR>> {{attachment:popupComputeSources.gif}} --- {{attachment:panelComputeSources.gif}} 1. With this first window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation. * Edit the Comment field if you want. * Select "Min. Norm Imaging" * For the sensors, you have no other option than MEG, because those recordings were recorded with only one type of MEG sensors (axial gradiometers) and no EEG signal was registrated at the same time. * Run. 1. The next window shows the options for the minimum norm estimation algorithm.<<BR>><<BR>> {{attachment:minNormOptions.gif}} * Leave the ''Tikhonov ''and ''Forward field normalization'' selected. To learn more about those options, please consult the the [[Theory]] section. * The ''Noise normalization'' option uses the noise covariance if it is available (else, use the signal variance) to normalize each channel by the level of noise in it during the source estimation. Leave this option checked. * However, you need to understand well what is the meaning of the last option, ''Output format'', which defines the way the sources are saved. * '''Full results''': Saves in one big matrix the values of all the sources (15000) for all the time samples (375). * The size in memory of such a matrix is about 45Mb for 300ms of recordings. This is still reasonable, so you may use this option in this case. * But if you need to process longer recordings, you may have some "Out of memory" errors in Matlab, or fill your hard drive quickly. * '''Kernel only''': Saves only the ''Inversion kernel'', a matrix that describes how to compute the sources when you know the values at the sensors level. So its size is: number of sources (15000) x number of sensors (151). * To get the sources time series, you just need to multiply this kernel by the MEG recordings. * '''Full results '''=''' Inversion kernel '''*''' Recordings''' * The size of this matrix is about 18Mb. In this case, the difference is not very important because we only process 375 time samples. But this inversion kernel is independent from the recordings length, so you can easily scale its computation to much longer recordings. * This is possible because the Minimum norm estimation is a linear method. * Which option should you choose ? * Probably "''Kernel only''", as it is faster and produces smaller files. * All the following operations in Brainstorm will be exactly the same whatever you select. Each time you will access the sources values, the program will have to do the multiplication Kernel * Recordings, but this is done in a totally transparent way. * The only reason that would make you chose the "''Full results''" options would be any interest in having the full matrix in one file, in case you want to process the sources values by yourself (filtering, statistics, display...). 1. So please check "''Kernel only''" for the moment. And click ''Ok''. |
1. Right-click on ''Subject01 / Right / ERF'' > ''Compute sources''.<<BR>><<BR>> {{attachment:popupComputeSources.gif}} --- {{attachment:panelComputeSources.gif}} 1. With this window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation. The default "Normal mode" only let you edit the following options:<<BR>> * '''Comment''': This field contains what is going to be displayed in the database explorer. * '''Method''': Please select wMNE. The other methods dSPM and sLORETA are also based on wMNE. They may give better and/or smoother results depending on the cases. * '''Sensors type''': Modalities that are used for the reconstruction. Here we only have one type of MEG sensors (axial gradiometers), so nothing to change. * '''Expert mode''': Show other options we do not care about right now. * Click on Run. |
| Line 37: | Line 19: |
| * It is displayed'' inside ''the recordings file ERP, because it is related to this file only. * You can have a look to what there is in the corresponding matrix file (right-click > File > View .mat file). You would find all the options of forward and inverse modelling, and only one interesting field : '''!ImagingKernel'''. |
* It is displayed'' inside ''the recordings file ERF, because it is related to this file only. * Meaning of that weird filename: "MN" stands for "Minimum Norm", and "Constr" stands for "Constrained orientation" of the dipoles (the estimated dipoles orientations are constrained to be normal to the cortex). * You can have a look to the corresponding matrix file (right-click > File > View file contents). You would find all the options of forward and inverse modeling, and only one interesting field : '''ImagingKernel''', which contains the inversion kernel. It is a [nVertices x nChannels] matrix that has to be multiplied with the recordings matrix in order to get the activity for each source at all the time samples. * The minimum norm solution being a linear operation (the time series for each source is a linear combination of all the time series recorded by the sensors), we make this economy of saving only this linear operator instead of the full source matrix (nVertices x nTime)<<BR>><<BR>> {{attachment:resultsMat.gif}} 1. Do the same for the ''Left / ERF'' file |
| Line 44: | Line 29: |
| 1. Double-click on recordings !StimRightThumb / ERP, to display the time series (always indispensable to have a time reference). 1. Double-click on sources !StimRightThumb / ERP / MN: MEG(Kernel). <<BR>>Equivalent to right-click > Cortical activations > Display on cortex. 1. Go to the main peak around 45ms (by clicking on the times series figure)<<BR>><<BR>> {{attachment:sources1.gif}} 1. Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recodings figures: rotation, zoom, ''Surfaces ''tab(smooth, curvature, resection...), colormap, sensors, pre-defined orientations (keys from 1 to 6)... 1. Only two new controls are available in the ''Surfaces ''tab, in panel ''Data options'': Threshold and transparency. * Transparency: changes the transparency of the sources on the cortex. * Threshold: Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed. |
1. Double-click on recordings ''Right / ERF'', to display the time series (always nice to have a time reference). 1. Double-click on sources ''Right / ERF / MN: MEG''. <<BR>>Equivalent to right-click > Cortical activations > Display on cortex. 1. Go to the main peak around 46ms (by clicking on the times series figure)<<BR>><<BR>> {{attachment:sources1.gif}} 1. Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recordings figures: rotation, zoom, ''Surface ''tab(smoothing, sulci, resection...), colormap, sensors, predefined orientations (keys from 0 to 7)... 1. Three new controls are available in the ''Surfaces ''tab, in panel ''Data options'': * '''Amplitude''': Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed. * '''Min. size''': Hide all the small activated regions, ie. the connected color patches that contain a number of vertices smaller than this "min.size" value. * '''Transparency''': Change the transparency of the sources on the cortex. |
| Line 52: | Line 38: |
| * The colorbar maximum depends on the way you configured your ''Sources ''colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 55 pA.m. * On the screenshot above, the threshold value was set to 34%. It means that only the sources that had a value over 0.34*55 = 18.7 pA.m were visible. |
* The colorbar maximum depends on the way you configured your ''Sources ''colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 68 pA.m. * On the screenshot above, the threshold value was set to 35%. It means that only the sources that had a value over 0.35*68 = 23.8 pA.m were visible. |
| Line 59: | Line 45: |
| 1. Close all the figures (''Close all'' button). Open the time series view for !StimRightThumb / ERP. 1. Right-click on !StimRightThumb / ERP / MN: MEG(Kernel) > Cortical activations > Display on MRI (3D). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.<<BR>><<BR>> {{attachment:sources3D.gif}} |
1. Close all the figures (''Close all'' button). Open the time series view for Right / ERF. 1. Right-click on Right / ERF / MN: MEG > Cortical activations > Display on MRI (3D). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.<<BR>><<BR>> {{attachment:sources3D.gif}} {{attachment:popupFigMri.gif}} 1. A new menu is available in the popup menu of this figure: MRI Display * '''MIP Anatomy''': for each slice, display the maximum value over all the slices instead of the original value in the structural MRI (fig 1) * '''MIP Functional''': same thing but with the layer of functional values (fig 2) * '''Smooth level''': The sources values are smoothed after being re-interpolated in the volume. These menus define the size of the smoothing kernel (fig2: smooth=2; fig3: smooth=5).<<BR>><<BR>> {{attachment:mriMipAnat.gif||height="143px",width="176px"}} {{attachment:mriMipFunc.gif||height="142px",width="175px"}} {{attachment:mriSmooth.gif||height="141px",width="173px"}} 1. This view can be used to lots of different types of contact sheets: in time or in space, for each orientation. You can try all the menus. Example: Right-click on the figure > Snapshot > Volume contact sheet: axial: <<BR>><<BR>> {{attachment:popupSnapshot.gif}} {{attachment:contactAxial.gif||height="288px",width="322px"}} |
| Line 64: | Line 55: |
| 1. Right-click on !StimRightThumb / ERP / MN: MEG(Kernel) > Cortical activations > Display on MRI (MRI Viewer). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to move the slices (sliders, mouse wheel, click on the views), move in time, change the threshold.<<BR>><<BR>> {{attachment:sourcesMriViwer.gif}} |
1. Right-click on Right / ERF / MN: MEG > Cortical activations > Display on MRI (MRI Viewer). 1. This view was also introduced in the tutorial about MRI and surface visualization. Try to move the slices (sliders, mouse wheel, click on the views), move in time, change the threshold.<<BR>><<BR>> {{attachment:sourcesMriViwer.gif||height="331px",width="359px"}} |
| Line 67: | Line 58: |
| === Modelized data === What is called ''modelized data ''here is the part of the recordings that are explained by the minimum-norm solution. This information is saved the sources file, in the ''Fsynth ''field. |
=== Minimum norm values are not only positive === You should pay attention to a property of the current amplitudes that are given by the wMNE method: they can be positive of negative, and they oscillate around zero. It's not easy to figure out what is the exact meaning of a negative value respect with a positive value, and most of the time we are only interested in knowing what is activated at what time, and therefore we look only at the absolute values of the sources. |
| Line 70: | Line 61: |
| 1. Close all the figures. Open the'' 2D sensor cap'' view for !StimRightThumb / ERP. 1. Right-click on ERP sources file > Modelized data > Display time series 1. Right-click on ERP sources file > Modelized data > 2D sensor cap 1. You can browse among time to observe what are the differences between what the model explains and the real recordings. The modelized data is smoother (right) that the original recordings (left): the regularization which is applied to obtain the sources is supposed to lower the noise.<<BR>><<BR>>''' {{attachment:topoReal.gif}} {{attachment:topoModelized.gif}} ''' |
In some other cases, mainly when doing frequency analysis, we need to pay attention to the sign of these values. Because we cannot do a frequency decomposition of the absolute values of the sources, we need to keep the sign all along our processes. |
| Line 75: | Line 63: |
| === Residuals === The ''residuals ''are the difference between the recordings and the modelized data. This information is not stored in any file but reconstructed when needed. |
Display again the sources for Right / ERF on the cortex surface (double-click on the source file), and uncheck the Absolute option for the colormap "Sources" (right-click on the figure > Colormap Sources > Absolute values). Decrease the threshold to observe the pattern of alternance between positive and negative values on the surface. Then double click on the colorbar to reset it to its default. |
| Line 78: | Line 65: |
| 1. Right-click on ERP sources > Residuals > Display time series<<BR>>At each time: Residuals = || Modelized - Recordings || / || max(Recordings) 1. Right-click on ERP sources > Residuals > 2D sensor cap: Spatial representation of the same values. 1. Right-click on ERP sources > Residuals > Display global error:<<BR>>At each time: Norm of residuals across all the sensors / Norm of the recordings across all the sensors<<BR>><<BR>>''' {{attachment:residualsTS.gif|residualsTopo.gif|height="163",width="216"}} {{attachment:residualsTopo.gif||height="162",width="212"}} {{attachment:residualsGlobal.gif||height="162",width="215"}} ''' |
{{attachment:relValues.gif}} === Minimum norm units === For information about the units used to represent the minimum norm source activation, pA.m (pico Ampere meter), please refer to the following forum post: [[http://neuroimage.usc.edu/forums/showthread.php?1246-Doubt-about-current-density-units-pA.m-or-pA-m2|http://neuroimage.usc.edu/...Doubt-about-current-density-units-pA.m-or-pA-m2]] |
| Line 83: | Line 73: |
| The sources file we are observing was computed as an ''inversion kernel''. It means that we can apply it to any similar recordings file (same subject, same run, same positions of sensors). But in our TutorialCTF database, the ''MN: MEG(Kernel)'' node only appears in the the ''ERP ''file, not in the ''Std ''one. What is nessary to share an inversion kernel between different recordings ? | The sources file we are observing was computed as an ''inversion kernel''. It means that we can apply it to any similar recordings file (same subject, same run, same positions of sensors). But in our TutorialCTF database, the ''MN: MEG'' node only appears in the the ''ERF ''file, not in the ''Std ''one. What is it necessary to share an inversion kernel between different recordings ? |
| Line 85: | Line 75: |
| 1. Compute another source estimation: but instead of clicking on the ''Compute sources'' fron the ''ERP ''recordings popup menu (which would mean that you only want sources for this particular recordings file), get this menu from the ''!StimRightThumb'' condition. This means that you want the inversion model to be applied to all the data in the condition.<<BR>><<BR>> {{attachment:popupComputeMulti.gif}} | 1. Compute another source estimation: but instead of clicking on the ''Compute sources'' from the ''ERF ''recordings popup menu (which would mean that you only want sources for this particular recordings file), get this menu from the ''Right''''' condition'''. This means that you want the inversion model to be applied to all the data in the condition. |
| Line 87: | Line 77: |
| 1. Select "Min. Norm Imaging", click Run, and then click Ok to validate the default options for minimum norm estimation. | 1. Select "Minimum Norm Imaging", click on Run. |
| Line 89: | Line 79: |
| 1. Three new nodes are available in the tree:<<BR>><<BR>> {{attachment:treeMinNormMulti.gif}} | 1. Three new nodes are available in the tree:<<BR>><<BR>> {{attachment:popupComputeMulti.gif}} {{attachment:treeMinNormMulti.gif}} |
| Line 91: | Line 81: |
| 1. The actual inversion kernel you have just computed (1), which contains the same information as the one from the previous section (Computing sources for a single data file). Note that you cannot do anything with this file: if you right-click on it, you will see that there are no ''Display ''menu for it. * Two links (2 and 3) that allow you apply this inversion kernel to the data files available in this condition (''ERP ''and ''Std''). In their popup menus, there are all the display options introduced in the previous section. |
1. The actual inversion kernel you have just computed (1), contains the same information as the one from the previous section (Computing sources for a single data file). Note that you cannot do anything with this file: if you right-click on it, you can see that there are no ''Display ''menu for it. * Two links (2) that allow you apply this inversion kernel to the data files available in this condition (''ERF ''and ''Std''). In their popup menus, there are all the display options introduced in the previous section. |
| Line 94: | Line 84: |
| * Right-click on the link in ERP (2) > File > View .mat file. The fields points to the recordings file (!DataFile) and the inversion kernel file (!LinkToFile). When you request any operation on this file, both recordings and kernel are loaded, multiplied to get the sources values, and then processed. <<BR>><<BR>> {{attachment:linkresultsMat.gif}} * The sources for the ''Std ''file do not have any meaning (it contains information about the averaging); but it illustrates the way a kernel is shared |
* These links are not saved as files but as specific strings in the database: "link|kernel_file|data_file". This means that to represent them, one should load the shared kernel, load the recordings, and multiply them. * The sources for the ''Std ''file do not have any meaning, do not even try to open it. It was just to illustrate the way a kernel is shared |
| Line 97: | Line 87: |
| 1. Double-click on both sources files available for StimRightThumb/ERP, and check at many different times that the cortical maps are exactly the same in both cases. | 1. Double-click on both sources files available for ''Right / ERF'' (link and non-link), and verify at many different times that the cortical maps are exactly the same in both cases. |
| Line 102: | Line 92: |
| == Minimum norm options == Let's introduce briefly the other options offered for the source estimation. Right-click again on Right / ERF > Compute sources. Click on "Expert mode", you see more options appearing in the window. If you click on Run, you have access to all the options of the wMNE algorithm. {{attachment:panelExpert.gif}} {{attachment:panelOptions.gif}} === Method === * '''wMNE''': Whitened and depth-weigthed linear L2-minimum norm estimates algorithm inspired from Matti Hamalainen's MNE software. Introduced in Brainstorm 3.1 by Rey Ramirez. For a full description of this method, please refer to the MNE manual, section 6, "The current estimates". [[http://www.nmr.mgh.harvard.edu/meg/manuals/MNE-manual-2.7.pdf|Download MNE manual here]]. * '''dSPM''': Noise-normalized estimate (dynamical Statistical Parametric Mapping [Dale, 2000]). Its computation is based on the wMNE solution. <<BR>>Basically, the dSPM value at each location is equal to the wMNE value divided by the projection of the estimated noise covariance matrix onto each source point. After whitening, the operational noise covariance matrix is by definition the identity matrix, and hence the projection of the noise is equal to the L2 norm of the row vector of the wMNE inverse operator (in the case of fixed dipole orientations). So, dSPM is what you get when the rows of the wMNE inverse operator all have unit norm (i.e., they all point in different directions but lie in a unit hyper-sphere). * '''sLORETA''': Noise-normalized estimate using the sLORETA approach (standardized LOw Resolution brain Electromagnetic TomogrAphy [Pasqual-Marqui, 2002]). sLORETA solutions have in general a smaller location bias than either the expected current (wMNE) or the dSPM. === Output mode === * '''Full results''': Saves in one big matrix the values of all the sources (15000) for all the time samples (375). The size in memory of such a matrix is about 45Mb for 300ms of recordings. This is still reasonable, so you may use this option in this case. But if you need to process longer recordings, you may have some "Out of memory" errors in Matlab, or fill your hard drive quickly. * '''Kernel only''': Saves only the ''Inversion kernel'', a matrix that describes how to compute the sources when you know the values at the sensor level. So its size is: number of sources (15000) x number of sensors (151). This is possible because these minimum norm methods are linear methods. * To get the sources time series, you just need to multiply this kernel by the MEG recordings. * '''Full results '''=''' Inversion kernel '''*''' Recordings''' * The size of this matrix is about 18Mb. In this case, the difference is not very important because we only process 375 time samples. But this inversion kernel is independent from the recordings length, so you can easily scale its computation to much longer recordings. * '''Default ?''' * Probably "''Kernel only''", as it is faster and produces smaller files. * All the following operations in Brainstorm will be exactly the same either way. Each time you access the sources values, the program has to do the multiplication Kernel * Recordings, but this is done in a totally transparent way. * The only reason that would make you choose the "''Full results''" options would be any interest in having the full matrix in one file, in case you want to process the sources values by yourself (filtering, statistics, display...). === Source orientation === * '''Constrained''': We consider that at each vertex of the cortex surface, there is only one dipole, and that its orientation is the normal to the cortex surface at this point. * The size of the inverse operator is [nVertices x nChannel]. * This is based on the anatomical observation that in the cortex, the neurons are mainly organized in macro-columns that are perpendicular to the cortex surface. But it's hard to know if we can really rely on it at this level, for this algorithm: Is it the case everywhere in the cortex? Are we supposed to use the inner (grey matter/white matter) or the outer (grey/CSF) surface of the cortex? Can we really be that precise in terms of MRI/MEG registration? These are questions that do not have final answers yet. * A technical advantage of this method, it produces one value per vertex instead of three. As a consequence 1) the output size is smaller, 2) it's faster to compute and display, and 3) the results are much more intuitive to display because we don't have to think about how to combine three values in one on a cortical map in a 3D figure * '''Unconstrained''': At each vertex of the cortex surface, we define a base of three dipoles with orthogonal directions, and then we estimate the sources for the three orientations independently.The size of the inverse operator is [3*nVertices x nChannel]. * '''Loose''': A version of the "unconstrained" method that integrates a weak orientation constraint. It generates an inverse operator that is the same size as the unconstrained one, but that emphasizes the importance of the sources that have an orientation that is close to the normal to the cortex. The value associated with this option set how "loose" should be the orientation constrain (recommended values in MNE are between 0.1 and 0.6, --loose option). This is the default in MNE software. * '''Truncated''':An SVD of the gain matrix for each source point is used to remove the dipole component with least variance, which for the single sphere head model, corresponds to the radial silent component. Only useful when combining a spherical head model and sLORETA. * '''Default''': At the present time, the default in Brainstorm is the "constrained" option, but we will probably switch to the unconstrained model soon. Because 1) we are not exactly sure that the orientation constraint is 100% correct, 2) the unconstrained sources look smoother and nicer, 3) the computation and storage capacities of the average computer increased a lot since the 1990s, so we can now afford to multiply by three the size of all the data. But right now, there are still some issues to fix in the processing pipeline of the unconstrained sources. === Signal properties === * '''Signal to noise ratio (SNR)''': An estimate of the amplitude SNR of the recordings, as defined in MNE (--snr option in MNE), used to compute the regularization parameter (lambda^2 = 1/SNR^2). The default value is SNR = 3. Automatic selection of the regularization parameter is currently not supported. * '''PCA Whitening''': Parameter introduced by Rey Ramirez. For more information, see the code of bst_wmne function. === Noise covariance matrix === * '''Full noise covariance''': Use the full noise covariance matrix available in Brainstorm database. If the noise covariance file previously computed in is a diagonal matrix (as it is the case in this tutorial), this value is ignored, and the "diagonal noise covariance" option is used instead. * '''Diagonal noise covariance''': Discard the off-diagonal elements of the noise covariance matrix (assuming heteroscedastic uncorrelated noise). Corresponds in MNE to the --diagnoise option. * '''Regularize noise covariance''': Regularize the noise-covariance matrix by the given amount for each type of sensor individually (value is restricted to the range 0...1). For more information, please refer to the MNE manual, section 6.2.4 (options --magreg, --gradreg and --eegreg). === Depth weighting === The minimum-norm estimates have a bias towards superficial currents. This tendency can be alleviated by adjusting these parameters. To understand how to set these parameters, please refer to the MNE manual (options --depth, --weightexp and --weightlimit). === Do it yourself === Typically, the only options you want to change is the type of estimator that is computed (wMNE, dSPM, sLORETA) and the orientation of the dipoles (constrained, unconstrained). Don't try to play with the other options if you are not really comfortable with what they represent. Now, compute and compare the following combinations of options for Right / ERF: 1. '''wMNE, Kernel only, Constrained '''(already computed) * Check the contents of the file (right-click > File > View file contents): The inverse operator is saved in the field ImagingKernel [nVertices x nSensors] 1. '''wMNE, Full results, Constrained''': * Check the contents of the file: The full results matrix is saved in the field ImageGridAmp [nVertices x nTime] * Open solutions #1 and #2 at the same time and check visually at different time points that the results are exactly the same 1. '''wMNE, Kernel only, Unconstrained''': * Check the contents of the file: The inverse operator contains now the information for three dipoles (three orientations) per vertex, [3*nVertices, nSensors] * Open solutions #1 and #3 at the same time, and observe that the unconstrained solution is much smoother 1. '''dSPM, Kernel only, Unconstrained''' 1. '''sLORETA, Kernel only, Unconstrained''' * Open solutions #3 (wMNE), #4 (dSPM) and #5 (sLORETA), all unconstrained. * Notice that the units are different: wMNE values are in pAm, dSPM and sLORETA are in arbitrary units (never try to compare these values to anything but the exact same type of inverse solution) * Observe around 46ms the respective behavior of these three solutions: * 3) wMNE tends to highlight the top of the gyri and the superficial sources, * 4) dSPM tends to correct that behavior and may give higher values in deeper areas, * 5) sLORETA produces a very smooth solution where all the potentially activated area of the brain (given to the low spatial resolution of the source localization with MEG/EEG) is shown as connected, regardless of the depth of the sources. wMNE: constrained (kernel and full), and unconstrained {{attachment:compMNEconstr.gif||height="173px",width="193px"}} {{attachment:compMNEconstr.gif||height="171px",width="190px"}} {{attachment:compMNE.gif||height="170px",width="189px"}} dSPM, sLORETA: {{attachment:compSPM.gif||height="173px",width="192px"}} {{attachment:compLORETA.gif||height="172px",width="191px"}} Now delete all these files when you're done, and keep only the initial solution: wMNE, Constrained. == Additional discussions on the forum == * Imaging resolution kernels: http://neuroimage.usc.edu/forums/showthread.php?1298 * Spatial smoothing of sources: http://neuroimage.usc.edu/forums/showthread.php?1409 * Units for dSPM and sLORETA: [[http://neuroimage.usc.edu/forums/showthread.php?1535-Dipole-strength-units-for-dSPM-and-sLORETA|http://neuroimage.usc.edu/forums/showthread.php?1535]] * EEG reference: http://neuroimage.usc.edu/forums/showthread.php?1525#post6718 * Sign of the MNE values: http://neuroimage.usc.edu/forums/showthread.php?1649#post7014 * Residual ocular artifacts: http://neuroimage.usc.edu/forums/showthread.php?1272 |
|
| Line 103: | Line 174: |
| You can now observe the cortical maps estimated from a MEG evoked potential. This information is interesting in itself, but it is an image; it is difficult to compare it rigorously with other subjects or stimulations, or to test statistically some hypothesis. | You can now view the cortical maps estimated from a MEG somatosensory evoked field. This information is interesting in itself, but it is an image; it is difficult to compare it rigorously with other subjects or experimental conditions. The next tutorial describes how to create and study a [[Tutorials/TutScouts|region of interest]]. |
| Line 105: | Line 176: |
| The next tutorial describes how to create and study a [[Tutorials/TutScouts|region of interest]]. | <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/TutSourceEstimation)>> |
[WARNING: This page is part of the old tutorials, please refer to the new documentation]
Tutorial 8: Source estimation
Authors: Francois Tadel, John C Mosher, Richard Leahy, Sylvain Baillet
Now you have in your database a forward model matrix that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we really need is to build the inverse information: how to estimate the sources when you have the recordings. Many solutions are described in the literature, some of them are implemented in Brainstorm, and only one is presented in this tutorial: the minimum-norm estimation. It is not really the most advanced solution, but it is one of the most robust.
Contents
Computing sources for a single data file
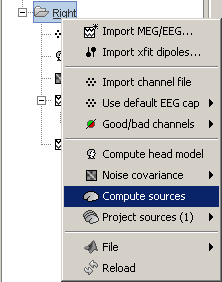
Right-click on Subject01 / Right / ERF > Compute sources.
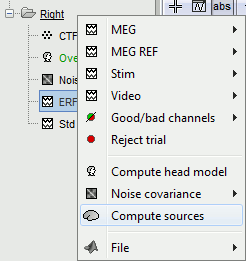 ---
--- 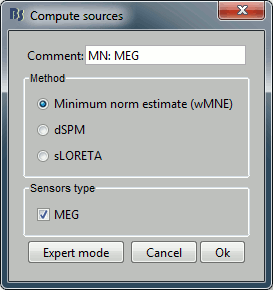
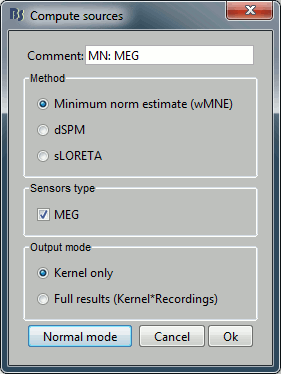
With this window you can select the method you want to use to estimate the cortical currents, and the sensors you are going to use for this estimation. The default "Normal mode" only let you edit the following options:
Comment: This field contains what is going to be displayed in the database explorer.
Method: Please select wMNE. The other methods dSPM and sLORETA are also based on wMNE. They may give better and/or smoother results depending on the cases.
Sensors type: Modalities that are used for the reconstruction. Here we only have one type of MEG sensors (axial gradiometers), so nothing to change.
Expert mode: Show other options we do not care about right now.
- Click on Run.
A new file is available in the database explorer.
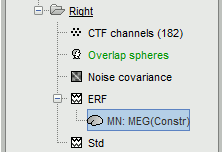
It is displayed inside the recordings file ERF, because it is related to this file only.
- Meaning of that weird filename: "MN" stands for "Minimum Norm", and "Constr" stands for "Constrained orientation" of the dipoles (the estimated dipoles orientations are constrained to be normal to the cortex).
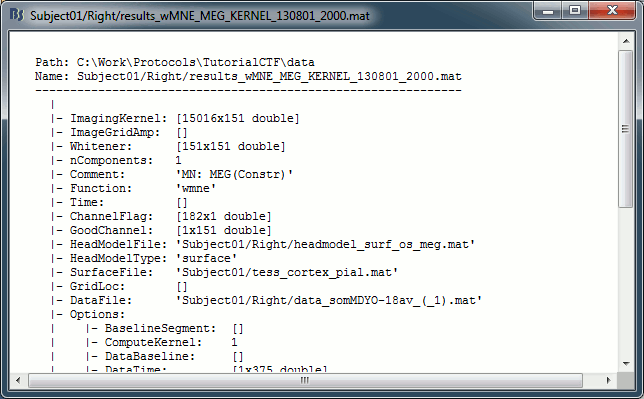
You can have a look to the corresponding matrix file (right-click > File > View file contents). You would find all the options of forward and inverse modeling, and only one interesting field : ImagingKernel, which contains the inversion kernel. It is a [nVertices x nChannels] matrix that has to be multiplied with the recordings matrix in order to get the activity for each source at all the time samples.
The minimum norm solution being a linear operation (the time series for each source is a linear combination of all the time series recorded by the sensors), we make this economy of saving only this linear operator instead of the full source matrix (nVertices x nTime)
Do the same for the Left / ERF file
Sources visualization
There are two main ways to display the sources: on the cortex surface and on the MRI slices.
Sources on cortex surface
Double-click on recordings Right / ERF, to display the time series (always nice to have a time reference).
Double-click on sources Right / ERF / MN: MEG.
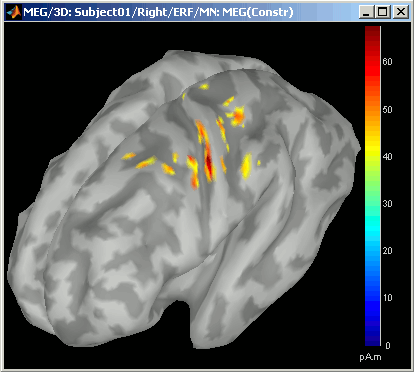
Equivalent to right-click > Cortical activations > Display on cortex.Go to the main peak around 46ms (by clicking on the times series figure)
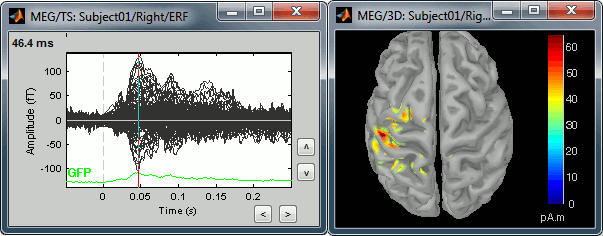
Then you can manipulate the sources display exactly the same way as the surfaces and the 2D/3D recordings figures: rotation, zoom, Surface tab(smoothing, sulci, resection...), colormap, sensors, predefined orientations (keys from 0 to 7)...
Three new controls are available in the Surfaces tab, in panel Data options:
Amplitude: Only the sources that have a value superior than a given percentage of the colorbar maximum are displayed.
Min. size: Hide all the small activated regions, ie. the connected color patches that contain a number of vertices smaller than this "min.size" value.
Transparency: Change the transparency of the sources on the cortex.
Take a few minutes to understand what this threshold value represents.
The colorbar maximum depends on the way you configured your Sources colormap. In case the colormap is NOT normalized to current time frame, and the maximum is NOT set to a specific value, the colorbar maximum should be around 68 pA.m.
- On the screenshot above, the threshold value was set to 35%. It means that only the sources that had a value over 0.35*68 = 23.8 pA.m were visible.
- If you set the threshold to 0%, you display all the sources values on the cortex surface; and as most of the sources have values close to 0, the brain is mainly blue.
Move the slider and look for a threshold value that would give you a really focal source.The following figures represent the sources activations at t=46ms respectively with threshold at 0% and 90%.
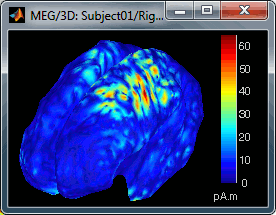
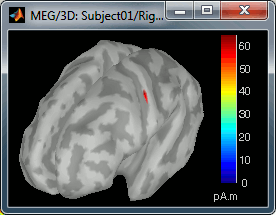
- The figure on the right shows the most active area of the cortex 46ms after an electric stimulation of the right thumb. As expected, it is localized in the left hemisphere, in the middle of post central gyrus (projection of the right hand in the primary somatosensory cortex).
Sources on MRI (3D)
Close all the figures (Close all button). Open the time series view for Right / ERF.
Right-click on Right / ERF / MN: MEG > Cortical activations > Display on MRI (3D).
This view was also introduced in the tutorial about MRI and surface visualization. Try to rotate, zoom, move the slices, move in time, change the threshold.
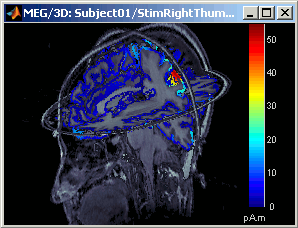
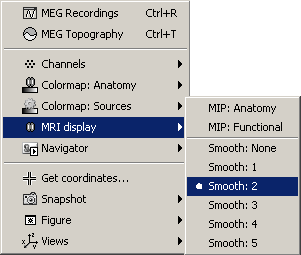
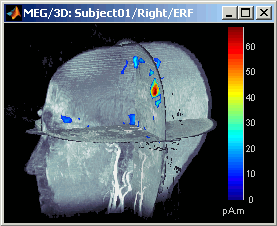
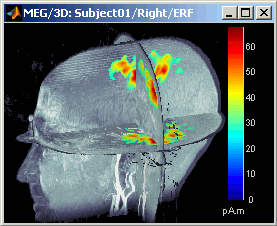
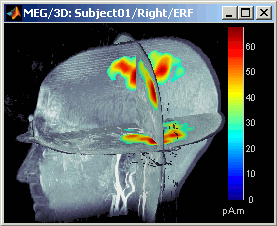
- A new menu is available in the popup menu of this figure: MRI Display
MIP Anatomy: for each slice, display the maximum value over all the slices instead of the original value in the structural MRI (fig 1)
MIP Functional: same thing but with the layer of functional values (fig 2)
Smooth level: The sources values are smoothed after being re-interpolated in the volume. These menus define the size of the smoothing kernel (fig2: smooth=2; fig3: smooth=5).
This view can be used to lots of different types of contact sheets: in time or in space, for each orientation. You can try all the menus. Example: Right-click on the figure > Snapshot > Volume contact sheet: axial:
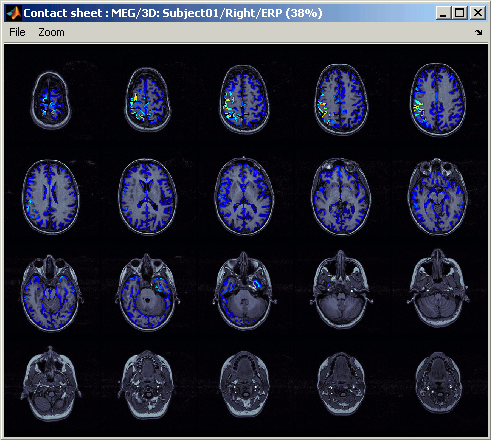
Sources on MRI (MRI Viewer)
Right-click on Right / ERF / MN: MEG > Cortical activations > Display on MRI (MRI Viewer).
This view was also introduced in the tutorial about MRI and surface visualization. Try to move the slices (sliders, mouse wheel, click on the views), move in time, change the threshold.
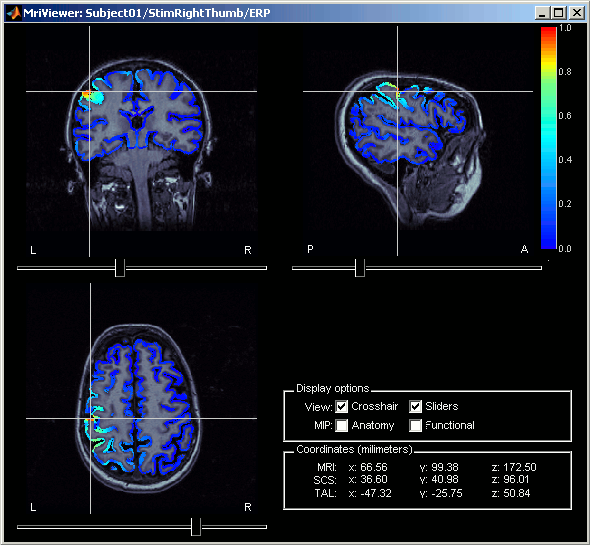
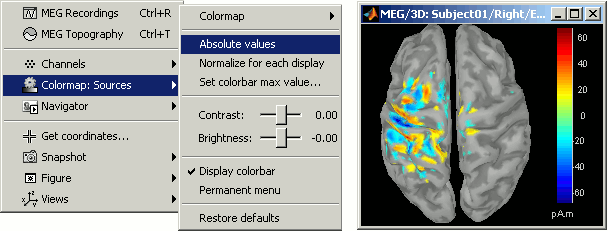
Minimum norm values are not only positive
You should pay attention to a property of the current amplitudes that are given by the wMNE method: they can be positive of negative, and they oscillate around zero. It's not easy to figure out what is the exact meaning of a negative value respect with a positive value, and most of the time we are only interested in knowing what is activated at what time, and therefore we look only at the absolute values of the sources.
In some other cases, mainly when doing frequency analysis, we need to pay attention to the sign of these values. Because we cannot do a frequency decomposition of the absolute values of the sources, we need to keep the sign all along our processes.
Display again the sources for Right / ERF on the cortex surface (double-click on the source file), and uncheck the Absolute option for the colormap "Sources" (right-click on the figure > Colormap Sources > Absolute values). Decrease the threshold to observe the pattern of alternance between positive and negative values on the surface. Then double click on the colorbar to reset it to its default.
Minimum norm units
For information about the units used to represent the minimum norm source activation, pA.m (pico Ampere meter), please refer to the following forum post:
http://neuroimage.usc.edu/...Doubt-about-current-density-units-pA.m-or-pA-m2
Computing sources for multiple data files
The sources file we are observing was computed as an inversion kernel. It means that we can apply it to any similar recordings file (same subject, same run, same positions of sensors). But in our TutorialCTF database, the MN: MEG node only appears in the the ERF file, not in the Std one. What is it necessary to share an inversion kernel between different recordings ?
Compute another source estimation: but instead of clicking on the Compute sources from the ERF recordings popup menu (which would mean that you only want sources for this particular recordings file), get this menu from the Right condition. This means that you want the inversion model to be applied to all the data in the condition.
- Select "Minimum Norm Imaging", click on Run.
Three new nodes are available in the tree:

The actual inversion kernel you have just computed (1), contains the same information as the one from the previous section (Computing sources for a single data file). Note that you cannot do anything with this file: if you right-click on it, you can see that there are no Display menu for it.
Two links (2) that allow you apply this inversion kernel to the data files available in this condition (ERF and Std). In their popup menus, there are all the display options introduced in the previous section.
- These links are not saved as files but as specific strings in the database: "link|kernel_file|data_file". This means that to represent them, one should load the shared kernel, load the recordings, and multiply them.
The sources for the Std file do not have any meaning, do not even try to open it. It was just to illustrate the way a kernel is shared
Double-click on both sources files available for Right / ERF (link and non-link), and verify at many different times that the cortical maps are exactly the same in both cases.
You can estimate the sources for many subjects or conditions at once, as it was explained for the head models in previous tutorial: the Compute sources menu is available on all the subjects and conditions popup menus.
- Delete the shared kernel (1), we don't need redundant and confusing information for the next steps. Observe that both links disappear at the same time.
Minimum norm options
Let's introduce briefly the other options offered for the source estimation. Right-click again on Right / ERF > Compute sources. Click on "Expert mode", you see more options appearing in the window. If you click on Run, you have access to all the options of the wMNE algorithm.
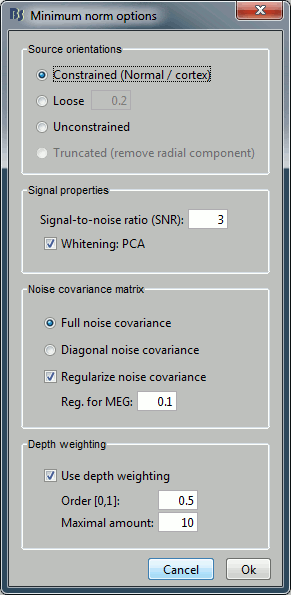
Method
wMNE: Whitened and depth-weigthed linear L2-minimum norm estimates algorithm inspired from Matti Hamalainen's MNE software. Introduced in Brainstorm 3.1 by Rey Ramirez. For a full description of this method, please refer to the MNE manual, section 6, "The current estimates". Download MNE manual here.
dSPM: Noise-normalized estimate (dynamical Statistical Parametric Mapping [Dale, 2000]). Its computation is based on the wMNE solution.
Basically, the dSPM value at each location is equal to the wMNE value divided by the projection of the estimated noise covariance matrix onto each source point. After whitening, the operational noise covariance matrix is by definition the identity matrix, and hence the projection of the noise is equal to the L2 norm of the row vector of the wMNE inverse operator (in the case of fixed dipole orientations). So, dSPM is what you get when the rows of the wMNE inverse operator all have unit norm (i.e., they all point in different directions but lie in a unit hyper-sphere).sLORETA: Noise-normalized estimate using the sLORETA approach (standardized LOw Resolution brain Electromagnetic TomogrAphy [Pasqual-Marqui, 2002]). sLORETA solutions have in general a smaller location bias than either the expected current (wMNE) or the dSPM.
Output mode
Full results: Saves in one big matrix the values of all the sources (15000) for all the time samples (375). The size in memory of such a matrix is about 45Mb for 300ms of recordings. This is still reasonable, so you may use this option in this case. But if you need to process longer recordings, you may have some "Out of memory" errors in Matlab, or fill your hard drive quickly.
Kernel only: Saves only the Inversion kernel, a matrix that describes how to compute the sources when you know the values at the sensor level. So its size is: number of sources (15000) x number of sensors (151). This is possible because these minimum norm methods are linear methods.
- To get the sources time series, you just need to multiply this kernel by the MEG recordings.
Full results = Inversion kernel * Recordings
- The size of this matrix is about 18Mb. In this case, the difference is not very important because we only process 375 time samples. But this inversion kernel is independent from the recordings length, so you can easily scale its computation to much longer recordings.
Default ?
Probably "Kernel only", as it is faster and produces smaller files.
- All the following operations in Brainstorm will be exactly the same either way. Each time you access the sources values, the program has to do the multiplication Kernel * Recordings, but this is done in a totally transparent way.
The only reason that would make you choose the "Full results" options would be any interest in having the full matrix in one file, in case you want to process the sources values by yourself (filtering, statistics, display...).
Source orientation
Constrained: We consider that at each vertex of the cortex surface, there is only one dipole, and that its orientation is the normal to the cortex surface at this point.
- The size of the inverse operator is [nVertices x nChannel].
- This is based on the anatomical observation that in the cortex, the neurons are mainly organized in macro-columns that are perpendicular to the cortex surface. But it's hard to know if we can really rely on it at this level, for this algorithm: Is it the case everywhere in the cortex? Are we supposed to use the inner (grey matter/white matter) or the outer (grey/CSF) surface of the cortex? Can we really be that precise in terms of MRI/MEG registration? These are questions that do not have final answers yet.
- A technical advantage of this method, it produces one value per vertex instead of three. As a consequence 1) the output size is smaller, 2) it's faster to compute and display, and 3) the results are much more intuitive to display because we don't have to think about how to combine three values in one on a cortical map in a 3D figure
Unconstrained: At each vertex of the cortex surface, we define a base of three dipoles with orthogonal directions, and then we estimate the sources for the three orientations independently.The size of the inverse operator is [3*nVertices x nChannel].
Loose: A version of the "unconstrained" method that integrates a weak orientation constraint. It generates an inverse operator that is the same size as the unconstrained one, but that emphasizes the importance of the sources that have an orientation that is close to the normal to the cortex. The value associated with this option set how "loose" should be the orientation constrain (recommended values in MNE are between 0.1 and 0.6, --loose option). This is the default in MNE software.
Truncated:An SVD of the gain matrix for each source point is used to remove the dipole component with least variance, which for the single sphere head model, corresponds to the radial silent component. Only useful when combining a spherical head model and sLORETA.
Default: At the present time, the default in Brainstorm is the "constrained" option, but we will probably switch to the unconstrained model soon. Because 1) we are not exactly sure that the orientation constraint is 100% correct, 2) the unconstrained sources look smoother and nicer, 3) the computation and storage capacities of the average computer increased a lot since the 1990s, so we can now afford to multiply by three the size of all the data. But right now, there are still some issues to fix in the processing pipeline of the unconstrained sources.
Signal properties
Signal to noise ratio (SNR): An estimate of the amplitude SNR of the recordings, as defined in MNE (--snr option in MNE), used to compute the regularization parameter (lambda2 = 1/SNR2). The default value is SNR = 3. Automatic selection of the regularization parameter is currently not supported.
PCA Whitening: Parameter introduced by Rey Ramirez. For more information, see the code of bst_wmne function.
Noise covariance matrix
Full noise covariance: Use the full noise covariance matrix available in Brainstorm database. If the noise covariance file previously computed in is a diagonal matrix (as it is the case in this tutorial), this value is ignored, and the "diagonal noise covariance" option is used instead.
Diagonal noise covariance: Discard the off-diagonal elements of the noise covariance matrix (assuming heteroscedastic uncorrelated noise). Corresponds in MNE to the --diagnoise option.
Regularize noise covariance: Regularize the noise-covariance matrix by the given amount for each type of sensor individually (value is restricted to the range 0...1). For more information, please refer to the MNE manual, section 6.2.4 (options --magreg, --gradreg and --eegreg).
Depth weighting
The minimum-norm estimates have a bias towards superficial currents. This tendency can be alleviated by adjusting these parameters. To understand how to set these parameters, please refer to the MNE manual (options --depth, --weightexp and --weightlimit).
Do it yourself
Typically, the only options you want to change is the type of estimator that is computed (wMNE, dSPM, sLORETA) and the orientation of the dipoles (constrained, unconstrained). Don't try to play with the other options if you are not really comfortable with what they represent. Now, compute and compare the following combinations of options for Right / ERF:
wMNE, Kernel only, Constrained (already computed)
Check the contents of the file (right-click > File > View file contents): The inverse operator is saved in the field ImagingKernel [nVertices x nSensors]
wMNE, Full results, Constrained:
Check the contents of the file: The full results matrix is saved in the field ImageGridAmp [nVertices x nTime]
- Open solutions #1 and #2 at the same time and check visually at different time points that the results are exactly the same
wMNE, Kernel only, Unconstrained:
- Check the contents of the file: The inverse operator contains now the information for three dipoles (three orientations) per vertex, [3*nVertices, nSensors]
- Open solutions #1 and #3 at the same time, and observe that the unconstrained solution is much smoother
dSPM, Kernel only, Unconstrained
sLORETA, Kernel only, Unconstrained
- Open solutions #3 (wMNE), #4 (dSPM) and #5 (sLORETA), all unconstrained.
- Notice that the units are different: wMNE values are in pAm, dSPM and sLORETA are in arbitrary units (never try to compare these values to anything but the exact same type of inverse solution)
- Observe around 46ms the respective behavior of these three solutions:
- 3) wMNE tends to highlight the top of the gyri and the superficial sources,
- 4) dSPM tends to correct that behavior and may give higher values in deeper areas,
- 5) sLORETA produces a very smooth solution where all the potentially activated area of the brain (given to the low spatial resolution of the source localization with MEG/EEG) is shown as connected, regardless of the depth of the sources.
wMNE: constrained (kernel and full), and unconstrained

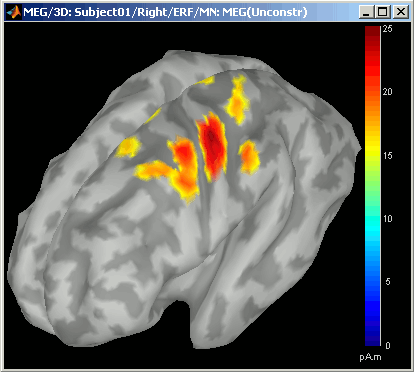
dSPM, sLORETA:
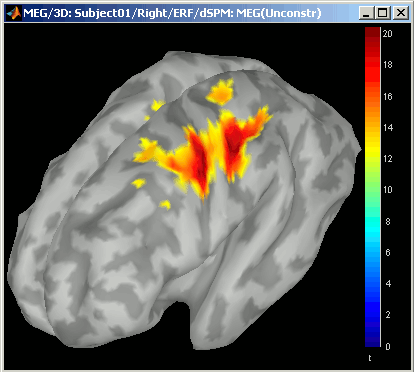
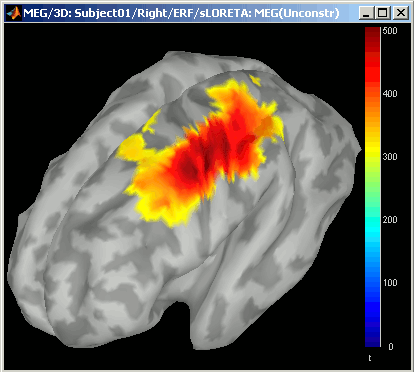
Now delete all these files when you're done, and keep only the initial solution: wMNE, Constrained.
Additional discussions on the forum
Imaging resolution kernels: http://neuroimage.usc.edu/forums/showthread.php?1298
Spatial smoothing of sources: http://neuroimage.usc.edu/forums/showthread.php?1409
Units for dSPM and sLORETA: http://neuroimage.usc.edu/forums/showthread.php?1535
EEG reference: http://neuroimage.usc.edu/forums/showthread.php?1525#post6718
Sign of the MNE values: http://neuroimage.usc.edu/forums/showthread.php?1649#post7014
Residual ocular artifacts: http://neuroimage.usc.edu/forums/showthread.php?1272
Next
You can now view the cortical maps estimated from a MEG somatosensory evoked field. This information is interesting in itself, but it is an image; it is difficult to compare it rigorously with other subjects or experimental conditions. The next tutorial describes how to create and study a ?region of interest.
