|
Size: 26427
Comment:
|
Size: 30397
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| ''Authors: Francois Tadel'' |
|
| Line 4: | Line 6: |
| If you are interested in running your own code from the Brainstorm interface and benefit from the powerful database and visualization systems, the best option is probably for you to create your own process functions. It can take some time to get used to this logic but it is time well invested: you will be able to exchange code easily with your collaborators and once your functions are stable, we can integrate them in the main Brainstorm distribution. The methods you develop can immediately reach thousands of users and we can maintain the code for you to ensure it stays compatible with the future releases of the software. | If you are interested in running your own code from the Brainstorm interface and benefit from the powerful database and visualization systems, the best option is probably for you to create your own process functions. It can take some time to get used to this logic but it is time well invested: you will be able to exchange code easily with your collaborators and the methods you develop could immediately reach thousands of users. Once your functions are stable, we can integrate them in the main Brainstorm distribution and maintain the code for you to ensure it stays compatible with the future releases of the software. |
| Line 8: | Line 10: |
| {{attachment:introPipeline.gif||height="283",width="489"}} | {{attachment:introPipeline.gif||width="489",height="283"}} <<TableOfContents(3,2)>> |
| Line 11: | Line 15: |
| Before you start reading this long technical reference, you should be aware of a simpler solution to execute your own code from the Brainstorm interface. The process "Pre-process > Run Matlab command" is very simple but very powerful. It loads the files in input and run the signals through a piece of Matlab code that you can edit freely. It can extend a lot the flexibility of the Brainstorm pipeline manager, providing an easy access to any Matlab function or script. |
Before you start reading this long technical document, you should be aware of a simpler solution to execute your own code from the Brainstorm interface. The process "Pre-process > '''Run Matlab command'''" is simple but very powerful. It loads the files in input and run them through a piece of Matlab code that you can edit freely. It can extend a lot the flexibility of the Brainstorm pipeline manager, providing an easy access to any Matlab function or script. |
| Line 21: | Line 25: |
| * '''$HOME/.brainstorm/process''':<<BR>>User processes folder, to develop new processes or overwrite some default function If you write a valid process function and place it in one of those folders, it will become automatically available in the pipeline editor menus, when you use the Process1 or Process2 tabs. Send it to another Brainstorm user and your code will be automatically available into the other person's Brainstorm interface. It is a very efficient way solution for exchanging code without the nightmare of understanding what are the inputs of the functions (units of the values, dimensions of the matrices, etc.). |
* '''$HOME/.brainstorm/process''':<<BR>>User folder, to develop new processes or overwrite Brainstorm functions If you write a new process function, name the script "process_...m" and place it in your '''user folder''', it will automatically become available in the pipeline editor menus, when you use the Process1 or Process2 tabs. Avoid using capital letters, spaces or special characters in the process file name.<<BR>>'''Warning''': Do not work directly in the brainstorm3 folder, or you will lose all your work the next time Brainstorm gets updated. Send it to another Brainstorm user and your code will automatically be available into the other person's Brainstorm interface. It is a very efficient solution for exchanging code without the nightmare of understanding what are the inputs of the functions (units of the values, dimensions of the matrices, etc.) |
| Line 31: | Line 35: |
| * '''process_test'''(): The first line of the script must contain a function with the same name as the .m script. It contains only a call to the Brainstorm script macro_methodcall. This allows us to call subfunction in the process_test.m script from outside, using the syntax: process_test('!FunctionName', arguments) * '''!GetDescription'''(): Returns a structure that describes the process: name, category, accepted inputs, options, etc. This function is called when Brainstorm parses the process folders to find all the valid processes. It informs the pipeline editor on how the process has to be integrated in the interface. * '''!FormatComment'''(): Returns a string that identifies the process in the interface. In the pipeline editor window, when the process is selected or when its options are modified, this function is called to update the process description line. Most processes would return simply the field sProcess.Comment, some other would add some options in the description (example: Pre-process > Band-pass filter, or Average > Average files). * '''Run'''(): Function called when the process is executed, either from the interface (after clicking on the Run button of the pipeline editor) or from a Matlab script (call to bst_process('!CallProcess', 'process_test', ...)). While the three first functions are descriptive, this one really does something. It receives the files placed in the Process1 or Process2 boxes, does its job and returns the output of the computation to Brainstorm. You are free to add as many sub-functions as needed to the process file. If your process needs some sub-functions to run, it is preferable to copy the full code directly into the "process_test.m" code, rather than leaving it in separate functions. This way it prevents from spreading subfunctions everywhere, which get later lost or forgotten in the distribution when the process is deleted. It might be uncomfortable at the beginning if you are not used to work with scripts with over 100 lines, but you'll get used to it, the Matlab code editor offers many solution to make long scripts easy to edit (cells, code folding...). It makes your process easier to maintain and to exchange with other users, which is important in the long run. |
* '''process_test'''(): The first line of the script must contain a function with the same name as the .m script. It contains only a call to the Brainstorm script macro_method. This allows to call sub-functions of the process_test.m script from other scripts, using the syntax: process_test('FunctionName', arguments) * '''GetDescription'''(): Returns a structure that describes the process: name, category, accepted inputs, options, etc. This function is called when Brainstorm parses the process folders to find all the valid processes. It informs the pipeline editor on how the process has to be integrated in the interface. * '''FormatComment'''(): Returns a string that identifies the process in the interface. In the pipeline editor window, when the process is selected or when its options are modified, this function is called to update the process description line. Most processes would return simply the field sProcess.Comment, some other would add some options in the description (example: Pre-process > Band-pass filter, or Average > Average files). * '''Run'''(): Function called when the process is executed, either from the interface (after clicking on the Run button of the pipeline editor) or from a Matlab script (call to bst_process('CallProcess', 'process_test', ...)). While the three first functions are descriptive, this one really does something. It receives the files placed in the Process1 or Process2 boxes, does its job and returns the output of the computation to Brainstorm. You are free to add as many sub-functions as needed to the process file. If your process needs some sub-functions to run, it is preferable to copy the full code directly into the "process_test.m" code, rather than leaving it in separate functions. This way it prevents from spreading sub-functions everywhere, which are later lost or forgotten in the distribution when the process is deleted. It might be uncomfortable at the beginning if you are not used to work with scripts with over 100 lines, but you'll get used to it, the Matlab code editor offers many solutions to make long scripts easy to edit (cells, code folding...). It makes your process easier to maintain and to exchange with other users, which is important in the long run. |
| Line 39: | Line 43: |
| Some processes can be designed to be called at the same time from the Brainstorm context, to work as a plug-in, and directly from the Matlab command line or a script, independently from the Brainstorm database and plug-in system. In this case, we can leave what is specific to the Brainstorm structure in the Run() function, and move the real computation to additional sub-functions. In this case, we recommend that you respect the following convention: name the main external sub-function '''Compute'''(). === Example: Z-score === Let's take the example of the process "Standardize > Z-score (static)", which is described in the function '''process_zscore.m'''. The function '''Run()''' reads and tests the options defined by the user and then calls Compute(), which is responsible from calculating the z-score normalization. |
A process can be designed to be called at the same time: * from the Brainstorm context, to work as a plug-in, and * from the Matlab command line or from a script, independently from the Brainstorm database. We can leave what is specific to the Brainstorm structure in the Run() function, and move the actual computation to additional sub-functions. In this case, we recommend that you respect the following convention: name the main external sub-function '''Compute'''(). The following example will help clarifying this concept. === Example: Notch filter === Let's take the example of the process "Pre-process > Notch filter", which is defined in the plug-in function '''brainstorm3/toolbox/process/functions/process_notch.m'''. The function '''GetDescription()''' defines the process properties (category, type of inputs, options...): {{{ function sProcess = GetDescription() % Description the process sProcess.Comment = 'Notch filter'; sProcess.Category = 'Filter'; [...] end }}} The function '''FormatComment()''' returns a string that represents the process in the interface: {{{ function Comment = FormatComment(sProcess) %#ok<DEFNU> if isempty(sProcess.options.freqlist.Value{1}) Comment = 'Notch filter: No frequency selected'; else strValue = sprintf('%1.0fHz ', sProcess.options.freqlist.Value{1}); Comment = ['Notch filter: ' strValue(1:end-1)]; end end }}} The function '''Run()''' reads and tests the options defined by the user and then calls Compute(): |
| Line 48: | Line 79: |
| % Get inputs iBaseline = panel_time('GetTimeIndices', sInput.TimeVector, sProcess.options.baseline.Value{1}); |
% Get options FreqList = sProcess.options.freqlist.Value{1}; |
| Line 51: | Line 82: |
| % Compute zscore sInput.A = Compute(sInput.A, iBaseline); |
% Filter data sInput.A = Compute(sInput.A, sfreq, FreqList); |
| Line 56: | Line 87: |
| The function '''Compute'''() calls another function '''!ComputeStat'''(): {{{ function A = Compute(A, iBaseline) % Calculate mean and standard deviation [meanBaseline, stdBaseline] = ComputeStat(A(:, iBaseline,:)); % Compute zscore A = bst_bsxfun(@minus, A, meanBaseline); A = bst_bsxfun(@rdivide, A, stdBaseline); end }}} {{{ function [meanBaseline, stdBaseline] = ComputeStat(A) % Compute baseline statistics stdBaseline = std(A, 0, 2); meanBaseline = mean(A, 2); % Remove null variance values stdBaseline(stdBaseline == 0) = 1e-12; end }}} This mechanism allows us to access this z-score function at different levels. We can call it as a Brainstorm process that takes Brainstorm structures in input (this is usually not done manually, but by the pipeline editor or by bst_process): |
The function '''Compute'''() applies a notch filter to the recordings in input: {{{ % USAGE: x = process_notch('Compute', x, sfreq, FreqList) function x = Compute(x, sfreq, FreqList) [...] % Remove the mean of the data before filtering xmean = mean(x,2); x = bst_bsxfun(@minus, x, xmean); % Remove all the frequencies sequencially [...] end }}} This mechanism allows us to access this notch filter at different levels. We can call it as a Brainstorm process that takes Brainstorm structures in input (this is usually not done manually): |
| Line 81: | Line 105: |
| Or as regular functions that take standard Matlab matrices in input: | As part of a script generated from the pipeline editor: {{{ % Process: Notch filter: 60Hz 120Hz 180Hz sFiles = bst_process('CallProcess', 'process_notch', sFiles, [], ... 'freqlist', [60, 120, 180], ... 'sensortypes', 'MEG, EEG', ... 'overwrite', 0); }}} Or as regular functions that takes standard Matlab matrices in input: |
| Line 85: | Line 118: |
| F = rand(1,500); ind = 1:100; % Normalize the signal F = process_zscore('Compute', F, ind); % Or just calculate its average and standard deviation [Favg, Fstd] = process_zscore('ComputeStat', F); |
sfreq = 1000; F = rand(1,10*sfreq); % Filter the signal F = process_notch('Compute', F, 1000, [60 120 180]); |
| Line 94: | Line 123: |
| The function !GetDescription() creates a structure sProcess that documents the process: its name, the way it is supposed to be used in the interface and all the options it needs. It contains the following fields: * '''Comment''': String that represents the process in the "Add process" menus of the pipeline editor window. * '''!FileTag''': String that is added to the description of the output files, in the case of "Filter" processes. In the example of the Z-score process, !FileTag='| zscore'. If you apply the Z-score process on a file named "MN: MEG", the file created by the process is named "MN: MEG | zscore". This file tag is also added to the file name. * '''Category''': String that defines how the process is supposed to behave. The possible values are defined in the next section: 'Filter', 'File', 'Custom'... * '''!SubGroup''': Sub-menu in which you want the process to appear in the menus of the pipeline editor. It can be an existing category (eg. 'Pre-processing', 'Standardize', etc) or a new category. |
The function GetDescription() creates a structure sProcess that documents the process: its name, the way it is supposed to be used in the interface and all the options it needs. It contains the following fields: * '''Comment''': String, represents the process in the "Add process" menus of the pipeline editor window. * '''FileTag''': String that is added to the description of the output files, in the case of "Filter" processes. In the example of the process "Absolute value", FileTag='abs'. If you apply this process on a file named "Avg: deviant", the file created by the process is named "Avg: deviant | abs". <<BR>>This file tag is also added to the file name. * '''Category''': String that defines how the process is supposed to behave.<<BR>>The possible values are defined in the next section: 'Filter', 'File', 'Custom'. * '''SubGroup''': Sub-menu in which you want the process to appear in the menus of the pipeline editor. It can be an existing category (eg. 'Pre-processing', 'Standardize', etc) or a new category. |
| Line 101: | Line 130: |
| * '''Description''': URL of the online tutorial describing the process. | |
| Line 102: | Line 132: |
| * '''!InputTypes''': Cell array of strings that represents the list of possible input types ('raw', 'data', 'results', 'timefreq', 'matrix'). This information is used to determine if a process is available in a specific interface configuration. For example: In the Process1 tab, if the "Process sources" button is selected, only the processes that have 'results' in their list !InputTypes, will marked as available. All the others will be greyed out. * '''!OutputTypes''': Cell array of strings with the same dimension as !InputTypes. It defines, for each input type, what is the type of the files in output. For example: a process that has !InputTypes={'data','results'} and !OutputTypes={'timefreq','timefreq'} transforms recordings in time-frequency objects, and sources in time-frequency objects. Now if !OutputTypes={'data','results'}, the type of the new files is the same as the one from the input file. |
* '''InputTypes''': Cell array of strings that represents the possible input types ('raw', 'data', 'results', 'timefreq', 'matrix'). This information is used to determine if a process is available in a specific interface configuration. For example: In the Process1 tab, if the "Process sources" button is selected, only the processes that have 'results' in their list InputTypes, will be marked as available. All the others will be greyed out. * '''OutputTypes''': Cell array of strings with the same dimension as InputTypes. It defines, for each input type, what is the type of the files in output. For example: a process that has InputTypes={'data','results'} and OutputTypes={'timefreq','timefreq'} transforms recordings in time-frequency objects, and sources in time-frequency objects. Now if OutputTypes={'data','results'}, the type of the new files is the same as the one from the input file. |
| Line 105: | Line 135: |
| * '''nMinFiles''': Integer, minimum number of files required by the process to run. Can be 1 or more. * '''processDim''': For the "Filter" processes, defines along which dimensions the process is allowed to split the input data matrix while processing it, if it's to big to be processed at once. Possible values: * 1: Split in blocks of signals (example: Band-pass filter or Sinusoid removal) * 2: Split in time blocks (example: EEG average reference, apply SSP) |
* '''nMinFiles''': Integer, minimum number of files required by the process to run (0, 1, 2 or more). * '''processDim''': For the "Filter" processes, defines along which dimensions the process is allowed to split the input data matrix while processing it, if it's too big to be processed at once. Possible values: * 1: Split in blocks of signals (example: Band-pass filter) * 2: Split in time blocks (example: EEG average reference or Apply SSP) |
| Line 110: | Line 140: |
| * '''isSourceAbsolute''': For the processes that accept source maps in input (type 'results'), this option defines if we want to process the real source values or their absolute values. The possible values are: | * '''isSourceAbsolute''': For the processes that accept source maps in input (type 'results'), this option defines if we want to process the real source values or their absolute values. Possible values: |
| Line 115: | Line 145: |
| * '''isPaired''': Applies only to the processes with two inputs (Process2), defines if the process needs pairs of files in input. If isPaired=1, the first file in FilesA and the first file in FilesB are processed together, and the second files are processed together, and so on. For example, it is the case for the paired t-test. * '''options''': List of options that are offered to the user in the pipeline editor window, and used by the !FormatComment() and Run() functions. This variable is a structure, where each field represents an option. Not all the fields have to be defined in the function !GetDescription(). The missing ones will be set to their default values, as defined in db_template('!ProcessDesc'). |
* '''isPaired''': Applies only to the processes with two inputs (Process2), defines if the process needs pairs of files in input. If isPaired=1, the first file in FilesA and the first file in FilesB are processed together, the second files are processed together and so on. Example: paired t-test. * '''options''': List of options that are offered to the user in the pipeline editor window. This variable is a structure, where each field represents an option. Not all the fields have to be defined in the function GetDescription(). The missing ones will be set to their default values, as defined in db_template('ProcessDesc'). |
| Line 125: | Line 155: |
| * '''Comment''': String, describes the option in the pipeline editor window (some option types ignore it) * '''Value''': Default value for this option. The type of this variable depends of the Type field * '''!InputTypes''': [optional] Cell array of the types of input files for which the option is shown |
* '''Comment''': String, describes the option in the pipeline editor window * '''Value''': Default value for this option. The type of this variable depends of the "Type" field * '''InputTypes''': [optional] Cell array of the types of input files for which the option is shown |
| Line 129: | Line 159: |
| * '''Class''': [optional] Groups options together to make them toggle-able (enabled or disabled) by a checkbox/radiobutton controller. Just give a common class name to all options to group together. * '''Controller''': [optional] * '''checkbox''': String (eg. 'Class1'), makes a checkbox type option control all options of class name specific in this field. * '''radio_linelabel''': struct('value1','Class1','value2','Class2'), makes the various options of a radio button group control other options in the same process. === Examples === |
|
| Line 149: | Line 184: |
| * ''''checkbox'''': A simple check box, to enable or disable something | * ''''checkbox'''': Simple check box, to enable or disable something |
| Line 152: | Line 187: |
| * Example: process_average * ''''radio'''': A list of radio buttons, to select between multiple choices * Comment: Cell array of strings, each string is a possible choice |
* Example: process_average.m * Note: Can also be used to enable or disable other options of the same Class with the Controller field. The disabled options will still have values, so you need to check in your code whether the checkbox has a Value of 1 or not to use these toggle-able options. * ''''radio'''': List of radio buttons, to select between multiple choices * Comment: Cell array of strings, each string is a possible choice ({comment1, comment2, ...}) |
| Line 156: | Line 192: |
| * Example: process_average * ''''combobox'''': A drop-down list, to select between multiple choice |
* Example: process_average.m * ''''radio_line'''': Same as 'radio', with all the options displayed on one single line * Comment: Cell array of strings, each string is a possible choice, with the comment of the line at the end: {comment1, comment2, comment3, ..., comment_line} * Value: Integer, index of the selected entry * Example: process_fft.m * ''''radio_label'''': Same as 'radio', with a label attached to each option (makes code easier to maintain) * Comment: {comment1, comment2, comment3; label1, label2, label3} * Value: String, label corresponding to the selected entry * Example: process_baseline.m * ''''radio_linelabel'''': Combination of 'radio_line' and 'radio_label' * Comment: {comment1, comment2, comment3, comment_line; label1, label2, label3, unused} * Value: String, label corresponding to the selected entry * Example: process_test_parametric2.m * ''''combobox'''': Drop-down list, to select between multiple choice |
| Line 160: | Line 208: |
| * Example: process_headmodel | * Example: process_headmodel.m |
| Line 164: | Line 212: |
| * Example: process_add_tag | * Example: process_add_tag.m |
| Line 168: | Line 216: |
| * Example: process_matlab_eval | * Example: process_matlab_eval.m |
| Line 175: | Line 223: |
| * Example: process_bandpass | * Example: process_bandpass.m |
| Line 179: | Line 227: |
| * [start,stop]: The two numerical values entered by the user | * [start,stop]: Two numerical values entered by the user |
| Line 182: | Line 230: |
| * Example: process_evt_detect * ''''timewindow'''': Same as 'range', but always offers by default the full time range covered by the first file in the files to process * Example: process_average_time * ''''baseline'''': Same as 'timewindow', but offers by default the time segment before 0 (or the full time range if there are no negative times) * ''''poststim'''': Same as 'timewindow', but offers by default the time segment after 0 (or the full time range if there are no negative times) |
* Example: process_evt_detect.m * ''''timewindow'''': Similar to 'range', but offers by default the time range of the first file to process * Example: process_average_time.m * ''''baseline'''': Same as 'timewindow', but offers by default the time segment before 0 * If there are no negative times, uses the full time range * ''''poststim'''': Same as 'timewindow', but offers by default the time segment after 0 * If there are no negative times, uses the full time range |
| Line 190: | Line 240: |
| * Example: process_average | * Example: process_average.m |
| Line 193: | Line 243: |
| * Value: !SelectOptions cell array, see example code * Example: process_evt_import * ''''datafile'''': Same as 'filename', but specific to MEG/EEG files that have to be imported or linked in the database * ''''channelname'''': Drop-down list to select a channel, can be edited directly by typing the channel name |
* Value: SelectOptions cell array, see details in the code of the example process * Example: process_evt_import.m * ''''datafile'''': Same as 'filename', but specific to MEG/EEG files * ''''channelname'''': Drop-down list to select a channel, can be edited directly |
| Line 199: | Line 249: |
| * Example: process_evt_detect * ''''subjectname'''': Drop-down list to select a subject, can be edited directly by typing the subject name |
* Example: process_evt_detect.m * ''''subjectname'''': Drop-down list to select a subject, can be edited directly |
| Line 203: | Line 253: |
| * Example: process_import_data_raw | * Example: process_import_data_raw.m |
| Line 207: | Line 257: |
| * Example: process_tf_bands * ''''cluster'''': Select a set of clusters or scouts in a list |
* Example: process_tf_bands.m * ''''cluster'''': Select a set of clusters in a list |
| Line 210: | Line 260: |
| * Value: Array of structures representing the scouts selected by the user * Example: process_extract_cluster * ''''cluster_confirm'''': Same as 'cluster', but with an additional checkbox on the top. If the checkbox is not selected by the user, the cluster/scout lists is greyed out and the returned Value will be []. * Example: process_fft * ''''atlas'''': Select from a drop-down list an atlas available in the the first input source file. The atlas selected by default in the list is the last one that one selected in the Scout tab with displaying the source file. |
* Value: Cell array with clusters selected by the user * Example: process_extract_cluster.m * ''''cluster_confirm'''': Same as 'cluster', but with an additional checkbox on the top * If the checkbox is not selected by the user, the cluster lists is greyed out and the returned Value will be []. * Example: TBD * ''''scout'''': Select a set of scouts in a list * Comment: Ignored * Value: Cell array with scouts selected by the user and the atlas they belong to. * Example: process_extract_scout.m * ''''scout_confirm'''': Same as 'scout', but with an additional checkbox on the top * If the checkbox is not selected by the user, the scout lists is greyed out and the returned Value will be []. * Example: process_extract_values.m * ''''atlas'''': Select from a drop-down list an atlas available in the the first input source file * The atlas selected by default in the list is the last one that one selected in the Scout tab with displaying the source file. |
| Line 217: | Line 276: |
| * Example: process_source_atlas | * Example: process_source_atlas.m |
| Line 220: | Line 279: |
| * Value: Structure returned by the function panel_function_name>!GetPanelContents() * Example: process_hilbert / panel_timefreq_options |
* Value: Structure returned by the function panel_function_name>GetPanelContents() * Example: process_hilbert.m / panel_timefreq_options.m * ''''button'''': Show a button that executes any piece of Matlab code when pressed * Comment: {'matlab_code', 'Comment', 'ButtonLabel'} * Value: Ignored * Example: process_bandpass.m |
| Line 224: | Line 287: |
| There are three different types of processes: '''Filter''', '''File''', '''Custom'''. The category of the process is defined by the field sProcess.Category. For the processes with two sets of inputs files (Process2), the logic is the same but the category are called: '''Filter2''', '''File2''', '''Custom'''. | There are three different types of processes: '''Filter''', '''File''', '''Custom'''. The category of the process is defined by the field sProcess.Category. For the processes with two sets of input files (Process2), the logic is the same but the category are called: '''Filter2''', '''File2''', '''Custom'''. |
| Line 227: | Line 290: |
| Brainstorm processes independently each file in the input list (the files that have been dropped in the Process1 or Process2 files lists) and is responsible for the following operations: | Brainstorm considers independently each file in the input list (the files that have been dropped in the Process1 or Process2 files lists) and is responsible for the following operations: |
| Line 242: | Line 305: |
| Limitations: There is no control over the file names and locations, one file in input = one file in output, and the file type cannot be changed (!InputTypes=!OutputTypes). | Limitations: There is no control over the file names and locations, one file in input = one file in output, and the file type cannot be changed (InputTypes=OutputTypes). Additionally, it is not possible to modify any field in the file other than the data matrix .F and the vector .Time. |
| Line 251: | Line 314: |
| The sInput structure gives lots of information about the input file coming from the database, and one additional fields "A" that contains the block of data to process. This process just applies the function abs() to the data sInput.A and returns modified values. A new file is created by Brainstorm in the database to store this result. | The sInput structure gives lots of information about the input file coming from the database, and one additional field "A" that contains the block of data to process. This process just applies the function abs() to the data sInput.A and returns modified values. A new file is created by Brainstorm in the database to store this result. |
| Line 254: | Line 317: |
| Brainstorm processes independently each file in the input list. It creates a structure sInput that documents the input file but does not load the data in the "A" field, as in the Filter case. | Brainstorm considers independently each file in the input list. It creates a structure sInput that documents the input file but does not load the data in the "A" field, as in the Filter case. |
| Line 261: | Line 324: |
| * referencing the new file in the database. [optional] * return the path of the new file |
* referencing the new file in the database, [optional] * returning the path of the new file. |
| Line 273: | Line 336: |
| OutputFile = bst_process('GetNewFilename', bst_fileparts(sStudy.FileName), fileType); | OutputFile=bst_process('GetNewFilename',bst_fileparts(sStudy.FileName),fileType); |
| Line 283: | Line 346: |
| The function Run() is called only once. It receives all the input file names in an array of structures "sInputs". From that, it can create zero, one or many files. The list of output files is returned in a cell array of strings "!OutputFiles". | The function Run() is called only once. It receives all the input file names in an array of structures "sInputs". It can create zero, one or many files. The list of output files is returned in a cell array of strings "OutputFiles". |
| Line 298: | Line 361: |
| * '''iItem''': Index of file in its category (example for recordings: sStudy.Data(sInput.iItem) * '''!FileName''': Input file name (file path relative to the protocol data folder) * '''!FileType''': Input file type ('raw', 'data', 'results', 'timefreq', 'matrix') |
* '''iItem''': Index of file in its category (example for recordings: sStudy.Data(sInput.iItem)) * '''FileName''': Input file name (file path relative to the data folder of the protocol) * '''FileType''': Input file type ('raw', 'data', 'results', 'timefreq', 'matrix') |
| Line 303: | Line 366: |
| * '''!SubjectFile''': Relative path to the subject file (brainstormsubject.mat) * '''!SubjectName''': Name of the subject * '''!DataFile''': For types 'results' or 'timefreq', path of the parent file in the database explorer * '''!ChannelFile''': Relative path to the channel file * '''!ChannelTypes''': Cell array of channel types available for the input file |
* '''SubjectFile''': Relative path to the subject file (brainstormsubject.mat) * '''SubjectName''': Name of the subject * '''DataFile''': For types 'results' or 'timefreq', path of the parent file in the database explorer * '''ChannelFile''': Relative path to the channel file * '''ChannelTypes''': Cell array of channel types available for the input file |
| Line 314: | Line 377: |
| * From a script generated by Brainstorm, using '''bst_process''': select the process in the pipeline editor window, select the menu "Generate .m script", then copy-paste the call to bst_process('!CallProcess', 'process_name', options). It can be executed from a script or directly from the Matlab command window * Directly calling a function within the process: with the syntax: "'''process_name'''('!FunctionName', arguments)" |
* From a script generated by Brainstorm, using '''bst_process''': select the process in the pipeline editor window, select the menu "Generate .m script", then copy-paste the call to bst_process. It can be executed from a script or directly from the Matlab command window.<<BR>> '''bst_process'''('CallProcess', 'process_name', options)<<BR>> * Directly calling a function within the process, with the syntax:<<BR>>'''process_name'''('FunctionName', arguments) |
| Line 318: | Line 381: |
| The easiest way for you to write your own plug-in function is to start working from an existing example. There are over a hundred processes currently available in the main Brainstorm distribution. Take some time to find one that is close to what you are planning to do. By order of importance: same category (Filter/File/Custom), same types of input and output files, similar options, same logic. Then follow those guidelines: | The easiest way for you to write your own plug-in function is to start working from an existing example. There are over a hundred processes currently available in the main Brainstorm distribution. Take some time to find one that is close to what you are planning to do. By order of importance: same category (Filter/File/Custom), same types of input and output files, similar options, same logic. Then follow these guidelines: |
| Line 321: | Line 384: |
| * Rename it so that it doesn't interfere with the existing functions: pick a name that represents what your function does and avoid using capital letters in the file name | * Rename it so that it doesn't interfere with the existing functions: pick a name that represents what your function does and avoid using capital letters, spaces or special characters in the file name |
| Line 324: | Line 387: |
| * Edit the some fields in the function !GetDescription functions: | * Edit some fields in the function GetDescription functions: |
| Line 326: | Line 389: |
| * sProcess.!SubGroup and sProcess.Index, so that it appears where you want in the list | * sProcess.SubGroup and sProcess.Index, so that it appears where you want in the list |
| Line 328: | Line 391: |
| * Edit the !FormatComment() function if needed. If it contains some complicated code, just replace it with "Comment = sProcess.Comment;" | * Edit the FormatComment() function if needed. If it contains some complicated code, just replace it with "Comment = sProcess.Comment;" |
| Line 332: | Line 395: |
| * Your process should be automatically available in the menus in the pipeline editor window. It is not, it might be because there are some errors in it: check your Matlab command window. If you see a message "Invalid plug-in function", Matlab could not execute the code correctly, check the syntax. | * Your process should be automatically available in the menus in the pipeline editor window. If it is not, it might be because there are some errors in it: check your Matlab command window. If you see a message "Invalid plug-in function", Matlab could not execute the code correctly, check the syntax. |
| Line 334: | Line 397: |
== Examples == Here are some additional sample processes that can help you for specific tasks: * Generate a head model / leadfield matrix: [[https://github.com/brainstorm-tools/bst-users/blob/master/processes/examples/process_headmodel_test.m|process_headmodel_test.m]] * Generate an inverse model / source matrix: [[https://github.com/brainstorm-tools/bst-users/blob/master/processes/examples/process_beamformer_test.m|process_beamformer_test.m]] * Load all the trials in input at once and process them: [[https://github.com/brainstorm-tools/bst-users/blob/master/processes/examples/process_example_customavg.m|process_example_customavg.m]] <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/TutUserProcess)>> |
How to write your own process
Authors: Francois Tadel
Brainstorm offers a flexible plug-in structure. All the operations available when using the Process1 and Process2 tabs, which means most of the Brainstorm features, are in fact written as plug-ins.
If you are interested in running your own code from the Brainstorm interface and benefit from the powerful database and visualization systems, the best option is probably for you to create your own process functions. It can take some time to get used to this logic but it is time well invested: you will be able to exchange code easily with your collaborators and the methods you develop could immediately reach thousands of users. Once your functions are stable, we can integrate them in the main Brainstorm distribution and maintain the code for you to ensure it stays compatible with the future releases of the software.
This tutorial looks long and complicated, but don't let it scare you. Putting your code in a process is not so difficult. Most of it is a reference manual that details all the possible options, you don't need to understand it completely. The last part explains how to copy an existing process and modify it to do what you want.
Contents
Alternative
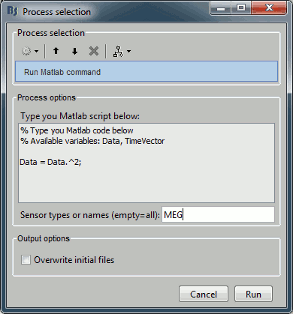
Before you start reading this long technical document, you should be aware of a simpler solution to execute your own code from the Brainstorm interface.
The process "Pre-process > Run Matlab command" is simple but very powerful. It loads the files in input and run them through a piece of Matlab code that you can edit freely. It can extend a lot the flexibility of the Brainstorm pipeline manager, providing an easy access to any Matlab function or script.
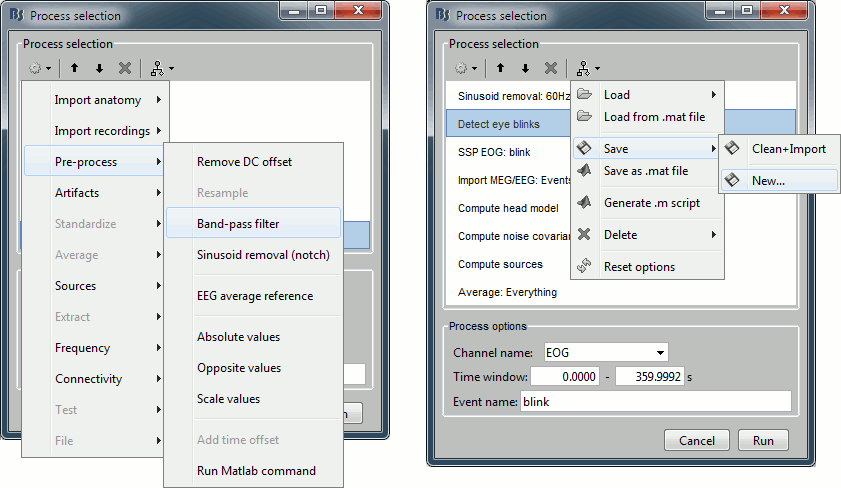
Process folders
A Brainstorm plug-in, or "process", is a single Matlab .m script that is automatically identified and added to the menus in the pipeline editor. Two folders are parsed for plug-ins:
brainstorm3/toolbox/process/functions:
Brainstorm "official" processes, included in the main distribution of the software$HOME/.brainstorm/process:
User folder, to develop new processes or overwrite Brainstorm functions
If you write a new process function, name the script "process_...m" and place it in your user folder, it will automatically become available in the pipeline editor menus, when you use the Process1 or Process2 tabs. Avoid using capital letters, spaces or special characters in the process file name.
Warning: Do not work directly in the brainstorm3 folder, or you will lose all your work the next time Brainstorm gets updated.
Send it to another Brainstorm user and your code will automatically be available into the other person's Brainstorm interface. It is a very efficient solution for exchanging code without the nightmare of understanding what are the inputs of the functions (units of the values, dimensions of the matrices, etc.)
Structure of the process scripts
Sub-functions
A process function must be named "process_...m" and located in one of the two process folders in order to be recognized by the software. Let's call our example function "process_test.m". It contains at least 4 functions:
process_test(): The first line of the script must contain a function with the same name as the .m script. It contains only a call to the Brainstorm script macro_method. This allows to call sub-functions of the process_test.m script from other scripts, using the syntax: process_test('FunctionName', arguments)
GetDescription(): Returns a structure that describes the process: name, category, accepted inputs, options, etc. This function is called when Brainstorm parses the process folders to find all the valid processes. It informs the pipeline editor on how the process has to be integrated in the interface.
FormatComment(): Returns a string that identifies the process in the interface. In the pipeline editor window, when the process is selected or when its options are modified, this function is called to update the process description line. Most processes would return simply the field sProcess.Comment, some other would add some options in the description (example: Pre-process > Band-pass filter, or Average > Average files).
Run(): Function called when the process is executed, either from the interface (after clicking on the Run button of the pipeline editor) or from a Matlab script (call to bst_process('CallProcess', 'process_test', ...)). While the three first functions are descriptive, this one really does something. It receives the files placed in the Process1 or Process2 boxes, does its job and returns the output of the computation to Brainstorm.
You are free to add as many sub-functions as needed to the process file. If your process needs some sub-functions to run, it is preferable to copy the full code directly into the "process_test.m" code, rather than leaving it in separate functions. This way it prevents from spreading sub-functions everywhere, which are later lost or forgotten in the distribution when the process is deleted. It might be uncomfortable at the beginning if you are not used to work with scripts with over 100 lines, but you'll get used to it, the Matlab code editor offers many solutions to make long scripts easy to edit (cells, code folding...). It makes your process easier to maintain and to exchange with other users, which is important in the long run.
Optional function: Compute()
A process can be designed to be called at the same time:
- from the Brainstorm context, to work as a plug-in, and
- from the Matlab command line or from a script, independently from the Brainstorm database.
We can leave what is specific to the Brainstorm structure in the Run() function, and move the actual computation to additional sub-functions. In this case, we recommend that you respect the following convention: name the main external sub-function Compute(). The following example will help clarifying this concept.
Example: Notch filter
Let's take the example of the process "Pre-process > Notch filter", which is defined in the plug-in function brainstorm3/toolbox/process/functions/process_notch.m.
The function GetDescription() defines the process properties (category, type of inputs, options...):
function sProcess = GetDescription()
% Description the process
sProcess.Comment = 'Notch filter';
sProcess.Category = 'Filter';
[...]
endThe function FormatComment() returns a string that represents the process in the interface:
function Comment = FormatComment(sProcess) %#ok<DEFNU>
if isempty(sProcess.options.freqlist.Value{1})
Comment = 'Notch filter: No frequency selected';
else
strValue = sprintf('%1.0fHz ', sProcess.options.freqlist.Value{1});
Comment = ['Notch filter: ' strValue(1:end-1)];
end
endThe function Run() reads and tests the options defined by the user and then calls Compute():
function sInput = Run(sProcess, sInput)
% Get options
FreqList = sProcess.options.freqlist.Value{1};
[...]
% Filter data
sInput.A = Compute(sInput.A, sfreq, FreqList);
[...]
endThe function Compute() applies a notch filter to the recordings in input:
% USAGE: x = process_notch('Compute', x, sfreq, FreqList)
function x = Compute(x, sfreq, FreqList)
[...]
% Remove the mean of the data before filtering
xmean = mean(x,2);
x = bst_bsxfun(@minus, x, xmean);
% Remove all the frequencies sequencially
[...]
endThis mechanism allows us to access this notch filter at different levels. We can call it as a Brainstorm process that takes Brainstorm structures in input (this is usually not done manually):
sInput = process_zscore('Run', sProcess, sInput);As part of a script generated from the pipeline editor:
% Process: Notch filter: 60Hz 120Hz 180Hz
sFiles = bst_process('CallProcess', 'process_notch', sFiles, [], ...
'freqlist', [60, 120, 180], ...
'sensortypes', 'MEG, EEG', ...
'overwrite', 0);Or as regular functions that takes standard Matlab matrices in input:
% Generate some random signal
sfreq = 1000; F = rand(1,10*sfreq);
% Filter the signal
F = process_notch('Compute', F, 1000, [60 120 180]);
Process description
The function GetDescription() creates a structure sProcess that documents the process: its name, the way it is supposed to be used in the interface and all the options it needs. It contains the following fields:
Comment: String, represents the process in the "Add process" menus of the pipeline editor window.
FileTag: String that is added to the description of the output files, in the case of "Filter" processes. In the example of the process "Absolute value", FileTag='abs'. If you apply this process on a file named "Avg: deviant", the file created by the process is named "Avg: deviant | abs".
This file tag is also added to the file name.Category: String that defines how the process is supposed to behave.
The possible values are defined in the next section: 'Filter', 'File', 'Custom'.SubGroup: Sub-menu in which you want the process to appear in the menus of the pipeline editor. It can be an existing category (eg. 'Pre-processing', 'Standardize', etc) or a new category.
Index: Integer value that indicates a relative position in the "Add process" menus. For example, if your process sets Index=411, it would be displayed after the Z-score process (Index=410). Two processes can have the same index, in this case the one displayed first is the one that is read first in the process folder. If you set the Index to zero, it would be ignored and not displayed in the menus of the pipeline editor.
Description: URL of the online tutorial describing the process.
isSeparator: Display a separator bar after the process in the pipeline editor menus.
InputTypes: Cell array of strings that represents the possible input types ('raw', 'data', 'results', 'timefreq', 'matrix'). This information is used to determine if a process is available in a specific interface configuration. For example: In the Process1 tab, if the "Process sources" button is selected, only the processes that have 'results' in their list InputTypes, will be marked as available. All the others will be greyed out.
OutputTypes: Cell array of strings with the same dimension as InputTypes. It defines, for each input type, what is the type of the files in output. For example: a process that has InputTypes={'data','results'} and OutputTypes={'timefreq','timefreq'} transforms recordings in time-frequency objects, and sources in time-frequency objects. Now if OutputTypes={'data','results'}, the type of the new files is the same as the one from the input file.
nInputs: Integer, defines the number of inputs of the process. If nInputs=1, the process appears in the Process1 tab. If nInputs=2, the process appears in the Process2 tab.
nMinFiles: Integer, minimum number of files required by the process to run (0, 1, 2 or more).
processDim: For the "Filter" processes, defines along which dimensions the process is allowed to split the input data matrix while processing it, if it's too big to be processed at once. Possible values:
- 1: Split in blocks of signals (example: Band-pass filter)
- 2: Split in time blocks (example: EEG average reference or Apply SSP)
- Empty: Does not allow the process to split the input data matrix (default)
isSourceAbsolute: For the processes that accept source maps in input (type 'results'), this option defines if we want to process the real source values or their absolute values. Possible values:
- -1: Never process the absolute values of the sources
- 0: Offer it as an option, but disabled by default
- 1: Offer it as an option, and enables it by default
- 2: Always process the absolute values of the sources
isPaired: Applies only to the processes with two inputs (Process2), defines if the process needs pairs of files in input. If isPaired=1, the first file in FilesA and the first file in FilesB are processed together, the second files are processed together and so on. Example: paired t-test.
options: List of options that are offered to the user in the pipeline editor window. This variable is a structure, where each field represents an option.
Not all the fields have to be defined in the function GetDescription(). The missing ones will be set to their default values, as defined in db_template('ProcessDesc').
Definition of the options
Options structure
The field sProcess.options describes the list of options that are displayed in the pipeline editor window when the process is selected. It is a structure with one field per option. If we have an option named "overwrite", it is described in the structure sProcess.options.overwrite. Every option is a structure with the following fields:
Type: String, defines the type of option (checkbox, text field, etc.)
Comment: String, describes the option in the pipeline editor window
Value: Default value for this option. The type of this variable depends of the "Type" field
InputTypes: [optional] Cell array of the types of input files for which the option is shown
Hidden: [optional] If set to 1, the option is not displayed in the pipeline editor, but passed to the Run function in the sProcess structure. It can be a way to pass additional parameters to the Run function without overloading the user interface.
Class: [optional] Groups options together to make them toggle-able (enabled or disabled) by a checkbox/radiobutton controller. Just give a common class name to all options to group together.
Controller: [optional]
checkbox: String (eg. 'Class1'), makes a checkbox type option control all options of class name specific in this field.
radio_linelabel: struct('value1','Class1','value2','Class2'), makes the various options of a radio button group control other options in the same process.
Examples
Example of two options defined in process_zscore.m:
% === Baseline time window
sProcess.options.baseline.Comment = 'Baseline:';
sProcess.options.baseline.Type = 'baseline';
sProcess.options.baseline.Value = [];
% === Sensor types
sProcess.options.sensortypes.Comment = 'Sensor types or names (empty=all): ';
sProcess.options.sensortypes.Type = 'text';
sProcess.options.sensortypes.Value = 'MEG, EEG';
sProcess.options.sensortypes.InputTypes = {'data'};
User preferences
Note that the default values defined in sProcess.options are usually displayed only once. When the user modifies the option, the new value is saved in the user preferences and offered as the default the next time the process is selected in the pipeline editor.
If you modify the Value field in your process function, the default offered when you select the process in the pipeline editor may not change accordingly. This means that another default has been saved in the user preferences. To reset all the options to their real default values (as defined in the process functions), you can use the menu Reset options in the Pipeline menu of pipeline editor window.
Option types
'checkbox': Simple check box, to enable or disable something
- Comment: String, displayed next to the checkbox
- Value: 0 (not checked) or 1 (checked)
- Example: process_average.m
- Note: Can also be used to enable or disable other options of the same Class with the Controller field. The disabled options will still have values, so you need to check in your code whether the checkbox has a Value of 1 or not to use these toggle-able options.
'radio': List of radio buttons, to select between multiple choices
- Comment: Cell array of strings, each string is a possible choice ({comment1, comment2, ...})
- Value: Integer, index of the selected entry
- Example: process_average.m
'radio_line': Same as 'radio', with all the options displayed on one single line
- Comment: Cell array of strings, each string is a possible choice, with the comment of the line at the end: {comment1, comment2, comment3, ..., comment_line}
- Value: Integer, index of the selected entry
- Example: process_fft.m
'radio_label': Same as 'radio', with a label attached to each option (makes code easier to maintain)
- Comment: {comment1, comment2, comment3; label1, label2, label3}
- Value: String, label corresponding to the selected entry
- Example: process_baseline.m
'radio_linelabel': Combination of 'radio_line' and 'radio_label'
- Comment: {comment1, comment2, comment3, comment_line; label1, label2, label3, unused}
- Value: String, label corresponding to the selected entry
- Example: process_test_parametric2.m
'combobox': Drop-down list, to select between multiple choice
- Comment: String displayed before the drop-down list
- Value: {iSelected, {'entry1', 'entry2', ...}} (iSelected is the index of the selected entry)
- Example: process_headmodel.m
'text': Simple text field
- Comment: String displayed before the text field
- Value: String
- Example: process_add_tag.m
'textarea': Multi-line text editor
- Comment: String displayed above the text area
- Value: String
- Example: process_matlab_eval.m
'value': Text field to edit numerical values with fixed precision
- Comment: String displayed before the text field
- Value: {value, units, precision}
- value: Numerical value entered by the user
- units: String that represents the units, displayed after the field
- precision: Number of decimals after the point (0=integer)
- Example: process_bandpass.m
'range': Two text fields to enter an interval, [start, stop]
- Comment: String displayed before the text fields
- Value: {[start,stop], units, precision}
- [start,stop]: Two numerical values entered by the user
- units: String that represents the units, displayed after the two fields
- precision: Number of decimals after the point (0=integer)
- Example: process_evt_detect.m
'timewindow': Similar to 'range', but offers by default the time range of the first file to process
- Example: process_average_time.m
'baseline': Same as 'timewindow', but offers by default the time segment before 0
- If there are no negative times, uses the full time range
'poststim': Same as 'timewindow', but offers by default the time segment after 0
- If there are no negative times, uses the full time range
'label': Simple text label, no user input
- Comment: String, accepts HTML input
- Value: Ignored
- Example: process_average.m
'filename': Select a file or a folder (text box + button "...")
- Comment: String displayed before the text box
Value: SelectOptions cell array, see details in the code of the example process
- Example: process_evt_import.m
'datafile': Same as 'filename', but specific to MEG/EEG files
'channelname': Drop-down list to select a channel, can be edited directly
- Comment: String displayed before the drop-down list
- Value: String, name of the selected channel
- Example: process_evt_detect.m
'subjectname': Drop-down list to select a subject, can be edited directly
- Comment: String displayed before the drop-down list
- Value: String, name of the selected subject
- Example: process_import_data_raw.m
'groupbands': Edit a list of time or frequency bands with a text editor
- Comment: String displayed above the text area
Value: Cell array that describes the frequency band, one row per band:
{'band_name', 'band_range', 'band_function'}
The default frequency bands can be obtained with: bst_get('DefaultFreqBands')- Example: process_tf_bands.m
'cluster': Select a set of clusters in a list
- Comment: Ignored
- Value: Cell array with clusters selected by the user
- Example: process_extract_cluster.m
'cluster_confirm': Same as 'cluster', but with an additional checkbox on the top
- If the checkbox is not selected by the user, the cluster lists is greyed out and the returned Value will be [].
- Example: TBD
'scout': Select a set of scouts in a list
- Comment: Ignored
- Value: Cell array with scouts selected by the user and the atlas they belong to.
- Example: process_extract_scout.m
'scout_confirm': Same as 'scout', but with an additional checkbox on the top
- If the checkbox is not selected by the user, the scout lists is greyed out and the returned Value will be [].
- Example: process_extract_values.m
'atlas': Select from a drop-down list an atlas available in the the first input source file
- The atlas selected by default in the list is the last one that one selected in the Scout tab with displaying the source file.
- Comment: String displayed before the drop-down list
- Value: String, name of the atlas selected by the user
- Example: process_source_atlas.m
'editpref': Show a button 'Edit' that opens a user-defined option panel
- Comment: {'panel_function_name', 'Comment'}
Value: Structure returned by the function panel_function_name>GetPanelContents()
- Example: process_hilbert.m / panel_timefreq_options.m
'button': Show a button that executes any piece of Matlab code when pressed
Comment: {'matlab_code', 'Comment', 'ButtonLabel'}
- Value: Ignored
- Example: process_bandpass.m
Categories of process
There are three different types of processes: Filter, File, Custom. The category of the process is defined by the field sProcess.Category. For the processes with two sets of input files (Process2), the logic is the same but the category are called: Filter2, File2, Custom.
Category: 'Filter' and 'Filter2'
Brainstorm considers independently each file in the input list (the files that have been dropped in the Process1 or Process2 files lists) and is responsible for the following operations:
- reading the input files,
- possibly splitting them if they are too big,
- writing the output files on the hard drive,
- referencing them in the database.
In the process, the function Run():
- receives the data matrix to process, one file at a time,
- applies some operation on it,
- returns the processed values.
Advantages: All the complicated things are taken care of automatically, the functions can be very short.
Limitations: There is no control over the file names and locations, one file in input = one file in output, and the file type cannot be changed (InputTypes=OutputTypes). Additionally, it is not possible to modify any field in the file other than the data matrix .F and the vector .Time.
For example, let's consider one of the simplest processes: process_absolute.m. It just calculates the absolute value of the input data matrix. The Run() function is only one line long:
function sInput = Run(sProcess, sInput)
sInput.A = abs(sInput.A);
endThe sInput structure gives lots of information about the input file coming from the database, and one additional field "A" that contains the block of data to process. This process just applies the function abs() to the data sInput.A and returns modified values. A new file is created by Brainstorm in the database to store this result.
Category: 'File' and 'File2'
Brainstorm considers independently each file in the input list. It creates a structure sInput that documents the input file but does not load the data in the "A" field, as in the Filter case.
In the process, the function Run() is called once for each input file and is responsible for:
- reading the input file,
- processing it,
- saving the results in a new file on the hard drive, [optional]
- referencing the new file in the database, [optional]
- returning the path of the new file.
The resulting functions are much longer, but this time the process is free do anything, there are no restrictions. The outline of the typical Run() function can be described as following:
function OutputFile = Run(sProcess, sInput)
% Load input file
DataMat = in_bst_data(sInput.FileName);
% Apply some function to the data in DataMat
OutputMat = some_function(DataMat);
% Generate a new file name in the same folder
OutputFile=bst_process('GetNewFilename',bst_fileparts(sStudy.FileName),fileType);
% Save the new file
save(OutputFile, '-struct', 'OutputMat');
% Reference OutputFile in the database:
db_add_data(sInput.iStudy, OutputFile, OutputMat);
end
Category: 'Custom'
Similar to the previous case "File", but this time all the input files are passed at once to the process.
The function Run() is called only once. It receives all the input file names in an array of structures "sInputs". It can create zero, one or many files. The list of output files is returned in a cell array of strings "OutputFiles".
function OutputFiles = Run(sProcess, sInputs)
% Load input files
% Do something interesting
% Save new files
% Reference the new files in the database
% Return all the new file names in the cell-array OutputFiles
end
Input description
The structure sInput contains the following fields:
iStudy: Index of the study, get the structure with sStudy=bst_get('Study', sInput.iStudy)
iItem: Index of file in its category (example for recordings: sStudy.Data(sInput.iItem))
FileName: Input file name (file path relative to the data folder of the protocol)
FileType: Input file type ('raw', 'data', 'results', 'timefreq', 'matrix')
Comment: Comment field of the input file (what is displayed in the database explorer)
Condition: Name of the condition/folder in which the file is located
SubjectFile: Relative path to the subject file (brainstormsubject.mat)
SubjectName: Name of the subject
DataFile: For types 'results' or 'timefreq', path of the parent file in the database explorer
ChannelFile: Relative path to the channel file
ChannelTypes: Cell array of channel types available for the input file
A: Loaded data matrix, only in the case of a Filter process
Running a process
Three ways to run a process:
From the interface: select the process in the pipeline editor window, then click on the Run button.
From a script generated by Brainstorm, using bst_process: select the process in the pipeline editor window, select the menu "Generate .m script", then copy-paste the call to bst_process. It can be executed from a script or directly from the Matlab command window.
bst_process('CallProcess', 'process_name', options)
Directly calling a function within the process, with the syntax:
process_name('FunctionName', arguments)
Create your own process
The easiest way for you to write your own plug-in function is to start working from an existing example. There are over a hundred processes currently available in the main Brainstorm distribution. Take some time to find one that is close to what you are planning to do. By order of importance: same category (Filter/File/Custom), same types of input and output files, similar options, same logic. Then follow these guidelines:
- Copy the original process_...m script to your $HOME/.brainstorm/process folder
- Rename it so that it doesn't interfere with the existing functions: pick a name that represents what your function does and avoid using capital letters, spaces or special characters in the file name
- Edit the function name in the first line of the process, so that it matches the script file name
- Edit the comments in the header of the function
Edit some fields in the function GetDescription functions:
- sProcess.Comment: String that represents the process in the pipeline editor menu
sProcess.SubGroup and sProcess.Index, so that it appears where you want in the list
- sProcess.options: Read the above documentation
Edit the FormatComment() function if needed. If it contains some complicated code, just replace it with "Comment = sProcess.Comment;"
- Edit the Run() function
- Delete all the other sub-functions
- Post on the Brainstorm forums all the questions you need
- Your process should be automatically available in the menus in the pipeline editor window. If it is not, it might be because there are some errors in it: check your Matlab command window. If you see a message "Invalid plug-in function", Matlab could not execute the code correctly, check the syntax.
- For development/debugging purpose, remember that you don't have to use the Brainstorm interface to execute your code. It can be annoying to select hundreds of times per day your process from the pipeline editor.
Examples
Here are some additional sample processes that can help you for specific tasks:
Generate a head model / leadfield matrix: process_headmodel_test.m
Generate an inverse model / source matrix: process_beamformer_test.m
Load all the trials in input at once and process them: process_example_customavg.m
