MEG visual tutorial: Group analysis (BIDS)
[WORK IN PROGRESS: THIS TUTORIAL IS NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Elizabeth Bock.
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "Multimodal, Multisubject data fusion". It is part of a collective effort to document and standardize MEG/EEG group analysis, see Frontier's research topic: From raw MEG/EEG to publication: how to perform MEG/EEG group analysis with free academic software.
The data processed here consists in simultaneous MEG/EEG recordings of 16 subjects performing a simple visual task on a large number of famous, unfamiliar and scrambled faces. This tutorial follows another page that explains how to process one single subject in details.
Contents
- License
- Download and installation [TODO] [Update screen captures]
- Overview of the analysis
- Subject averages: Famous, Unfamiliar, Scrambled
- Subject averages: Faces
- Subject averages: Within-subject differences
- Subject averages: Filter and normalize
- Subject averages: Screen captures
- Group analysis: MEG/EEG
- Group analysis: Sources
- Scripting
License
This dataset was obtained from the OpenfMRI project (http://www.openfmri.org), accession #ds117. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset, Scientific Data (2015)
Any questions, please contact: rik.henson@mrc-cbu.cam.ac.uk
Download and installation [TODO] [Update screen captures]
First, make sure you have enough space on your hard drive, at least 40Gb:
The Brainstorm database with all the data imported, downloaded from this website: 20Gb
The same database after this tutorial: 40Gb
You can follow this tutorial after processing the recordings for the 16 good subjects (6 runs per subject) as illustrated in the single subject tutorial. Otherwise, we provide a Brainstorm protocol that includes all the imported data, downsampled at 275Hz:
Go to the Download page, download the file TutorialGroup.zip (20Gb).
- Unzip this file in your Brainstorm database folder (brainstorm_db).
In Brainstorm, menu File > Load protocol > Load from folder > Select brainstorm_db/TutorialGroup
The database you need in order to follow this tutorial should contain the following:
The individual anatomy imported from FreeSurfer for each subject (16 subjects).
- The sensor level averages (MEG+EEG) for each run (downsampled to 275Hz or not).
The forward model, noise covariance and inverse models for each subject and each run.
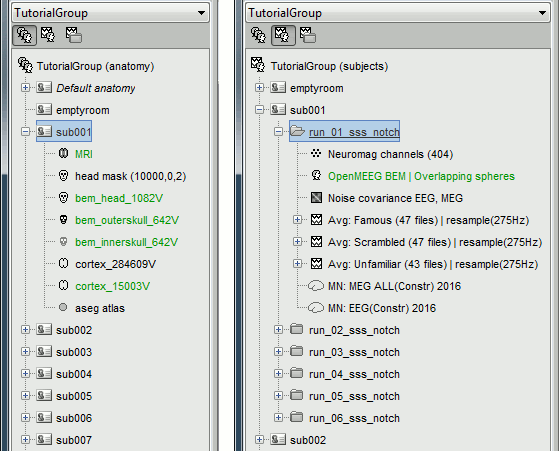
This protocol TutorialGroup is produced from the single subject protocol TutorialVisual with the script: brainstorm3/toolbox/script/tutorial_visual_copy.m
1 function tutorial_visual_copy(ProtocolNameSingle, ProtocolNameGroup, reports_dir) 2 % TUTORIAL_VISUAL_COPY: Copy the subject averages for the Brainstorm/SPM group tutorial into a new protocol (BIDS VERSION) 3 % 4 % ONLINE TUTORIALS: 5 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle 6 % - https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup 7 % 8 % INPUTS: 9 % - ProtocolNameSingle : Name of the protocol created with all the data imported (TutorialVisual) 10 % - ProtocolNameGroup : Name of the protocol with just the averages, downsampled to 275Hz (TutorialGroup) 11 % - reports_dir : If defined, exports all the reports as HTML to this folder 12 13 % @============================================================================= 14 % This function is part of the Brainstorm software: 15 % https://neuroimage.usc.edu/brainstorm 16 % 17 % Copyright (c) University of Southern California & McGill University 18 % This software is distributed under the terms of the GNU General Public License 19 % as published by the Free Software Foundation. Further details on the GPLv3 20 % license can be found at http://www.gnu.org/copyleft/gpl.html. 21 % 22 % FOR RESEARCH PURPOSES ONLY. THE SOFTWARE IS PROVIDED "AS IS," AND THE 23 % UNIVERSITY OF SOUTHERN CALIFORNIA AND ITS COLLABORATORS DO NOT MAKE ANY 24 % WARRANTY, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO WARRANTIES OF 25 % MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE, NOR DO THEY ASSUME ANY 26 % LIABILITY OR RESPONSIBILITY FOR THE USE OF THIS SOFTWARE. 27 % 28 % For more information type "brainstorm license" at command prompt. 29 % =============================================================================@ 30 % 31 % Author: Francois Tadel, Elizabeth Bock, 2016-2018 32 33 % ===== CHECK PROTOCOLS ===== 34 % Start brainstorm without the GUI 35 if ~brainstorm('status') 36 brainstorm nogui 37 end 38 % Output folder for reports 39 if (nargin < 3) || isempty(reports_dir) || ~isdir(reports_dir) 40 reports_dir = []; 41 end 42 % You have to specify the folder in which the tutorial dataset is unzipped 43 if (nargin < 2) || isempty(ProtocolNameSingle) || isempty(ProtocolNameGroup) 44 ProtocolNameSingle = 'TutorialVisual'; 45 ProtocolNameGroup = 'TutorialGroup'; 46 end 47 48 % Output protocol: Delete existing protocol 49 gui_brainstorm('DeleteProtocol', ProtocolNameGroup); 50 % Output protocol: Create new protocol 51 iProtocolGroup = gui_brainstorm('CreateProtocol', ProtocolNameGroup, 0, 0); 52 % Output protocol: Get protocol information 53 ProtocolInfoGroup = bst_get('ProtocolInfo'); 54 55 % Input protocol: Check that it exists 56 iProtocolSingle = bst_get('Protocol', ProtocolNameSingle); 57 if isempty(iProtocolSingle) 58 error(['Unknown protocol: ' ProtocolNameSingle]); 59 end 60 % Select input protocol 61 gui_brainstorm('SetCurrentProtocol', iProtocolSingle); 62 % Input protocol: Get protocol information 63 ProtocolInfoSingle = bst_get('ProtocolInfo'); 64 65 66 % ===== COPY ONLY GOOD SUBJECTS ===== 67 % Start a new report (one report per subject) 68 bst_report('Start'); 69 % Loop on subjects 70 for iSubj = 1:16 71 % Subject folders 72 SubjectName = sprintf('sub-%02d', iSubj); 73 AnatSrc = bst_fullfile(ProtocolInfoSingle.SUBJECTS, SubjectName); 74 DataSrc = bst_fullfile(ProtocolInfoSingle.STUDIES, SubjectName); 75 AnatDest = bst_fullfile(ProtocolInfoGroup.SUBJECTS, SubjectName); 76 DataDest = bst_fullfile(ProtocolInfoGroup.STUDIES, SubjectName); 77 % If subject folder doesn't exist: skip 78 if ~file_exist(AnatSrc) || ~file_exist(DataSrc) 79 disp(['Subject "' SubjectName '" does not exist or is incomplete.']); 80 continue; 81 end 82 % Copy anatomy files 83 mkdir(AnatDest); 84 disp(['Copying: ' AnatSrc ' to ' AnatDest '...']); 85 file_copy(bst_fullfile(AnatSrc, '*.mat'), AnatDest); 86 % Copy analysis folders 87 mkdir(bst_fullfile(DataDest, '@default_study')); 88 mkdir(bst_fullfile(DataDest, '@intra')); 89 disp(['Copying: ' DataSrc ' to ' DataDest '...']); 90 file_copy(bst_fullfile(DataSrc, '@default_study', '*.mat'), bst_fullfile(DataDest, '@default_study')); 91 file_copy(bst_fullfile(DataSrc, '@intra', '*.mat'), bst_fullfile(DataDest, '@intra')); 92 % Loop on runs 93 for iRun = 1:6 94 % Run folders 95 RunName = sprintf('sub-%02d_ses-meg_task-facerecognition_run-%02d_proc-sss_meg_notch', iSubj, iRun); 96 RunSrc = bst_fullfile(DataSrc, RunName); 97 RunDest = bst_fullfile(DataDest, RunName); 98 % If run folder doesn't exist: skip 99 if ~file_exist(RunSrc) 100 disp(['Run "' SubjectName '/' RunName '" does not exist or is incomplete.']); 101 continue; 102 end 103 % Copy files 104 mkdir(RunDest); 105 disp(['Copying: ' RunSrc ' to ' RunDest '...']); 106 file_copy(bst_fullfile(RunSrc, 'brainstormstudy.mat'), RunDest); 107 file_copy(bst_fullfile(RunSrc, 'channel_*.mat'), RunDest); 108 file_copy(bst_fullfile(RunSrc, '*_average_*.mat'), RunDest); 109 if ~isempty(dir(bst_fullfile(RunSrc, 'headmodel_*.mat'))) 110 file_copy(bst_fullfile(RunSrc, 'headmodel_*.mat'), RunDest); 111 end 112 if ~isempty(dir(bst_fullfile(RunSrc, 'noisecov_full.mat'))) 113 file_copy(bst_fullfile(RunSrc, 'noisecov_full.mat'), RunDest); 114 end 115 if ~isempty(dir(bst_fullfile(RunSrc, 'results_*.mat'))) 116 file_copy(bst_fullfile(RunSrc, 'results_*.mat'), RunDest); 117 end 118 if ~isempty(dir(bst_fullfile(RunSrc, 'timefreq_*.mat'))) 119 file_copy(bst_fullfile(RunSrc, 'timefreq_*.mat'), RunDest); 120 end 121 end 122 end 123 124 % ===== RELOAD ===== 125 % Reload output protocol 126 db_reload_database(iProtocolGroup); 127 % Select output protocol 128 gui_brainstorm('SetCurrentProtocol', iProtocolGroup); 129 130 % ===== DOWNSAMPLE TO 275HZ ===== 131 % Process: Select data files in: */* 132 sDataAll = bst_process('CallProcess', 'process_select_files_data', [], []); 133 % Process: Resample: 275Hz 134 sDataAll = bst_process('CallProcess', 'process_resample', sDataAll, [], ... 135 'freq', 275, ... 136 'overwrite', 1); 137 % Process: Select time-frequency files in: */* 138 sTimefreqAll = bst_process('CallProcess', 'process_select_files_timefreq', [], []); 139 % Process: Resample: 275Hz 140 if ~isempty(sTimefreqAll) 141 sTimefreqAll = bst_process('CallProcess', 'process_resample', sTimefreqAll, [], ... 142 'freq', 275, ... 143 'overwrite', 1); 144 end 145 146 % ===== RENAME: DATA ===== 147 % Process: Select data files in: */* 148 sDataAll = bst_process('CallProcess', 'process_select_files_data', [], []); 149 % Rename data files 150 for i = 1:length(sDataAll) 151 % Remove all the processing tags 152 iTag = strfind(sDataAll(i).Comment, ' |'); 153 if isempty(iTag) 154 continue; 155 end 156 newComment = sDataAll(i).Comment(1:iTag-1); 157 % Process: Set comment: AA 158 bst_process('CallProcess', 'process_set_comment', sDataAll(i), [], ... 159 'tag', newComment, ... 160 'isindex', 0); 161 end 162 163 % ===== RENAME: TIME-FREQ ===== 164 % Process: Select time-frequency files in: */* 165 sTimefreqAll = bst_process('CallProcess', 'process_select_files_timefreq', [], []); 166 % Rename timefreq files 167 %AllConditions = {'Famous', 'Scrambled', 'Unfamiliar'}; 168 for i = 1:length(sTimefreqAll) 169 % Remove all the processing tags 170 iTag = strfind(sTimefreqAll(i).Comment, ' |'); 171 if isempty(iTag) 172 continue; 173 end 174 newComment = sTimefreqAll(i).Comment(1:iTag-1); 175 %newComment = ['Avg: ', AllConditions{sTimefreqAll(i).iItem}, ', Power, 6-60Hz']; 176 % Process: Set comment 177 bst_process('CallProcess', 'process_set_comment', sTimefreqAll(i), [], ... 178 'tag', newComment, ... 179 'isindex', 0); 180 end 181 182 % Save report 183 ReportFile = bst_report('Save', []); 184 if ~isempty(reports_dir) && ~isempty(ReportFile) 185 bst_report('Export', ReportFile, bst_fullfile(reports_dir, ['report_' ProtocolNameGroup '_copy.html'])); 186 end 187 188 189 190 191
Overview of the analysis
Coregistration of the acquisition runs
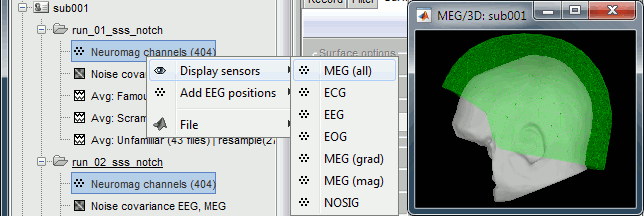
For each subject, all the runs have been registered to a common head position with MaxFilter. To verify this, select all the channel files within one subject, right-click > Display sensors > MEG (all). The surfaces representing the MEG helmet are perfectly overlapping for all the runs. When the runs are not aligned, it looks like this.
This means we can safely average or compare the MEG sensor values across runs within one subject. However, it is not reliable to average MEG recordings across subjects, because of the anatomical differences between subjects.
This doesn't mean we can estimate the sources only once per subject. We have computed different SSP projectors and selected different bad channels for each acquisition run. To be able to use this information efficiently we should estimate the sources for each run separately, then average the sources across runs.
The forward model is the same for all the runs within one subject, therefore it can be computed for the first run and copied to all the other runs.
Objectives
The objectives for this tutorial are to reproduce the analysis presented in the following documents:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset
Scientific Data (2015)Wakeman DG, Henson RN, Functional and structural connrctivity in face processing: MEG, EEG, fMRI, MRI and DWI data, Biomag 2016
SPM12 manual, chapter 41: Multimodal, Multisubject data fusion
Summary of the results we will compute in this tutorial:
- Subject and grand averages for each condition (Famous, Unfamiliar, Scrambled).
- Normalization of these averages (Z-score for the sources, or ERS/D for the time-frequency maps).
- Projection of the sources results on a template and spatial smoothing of the source maps.
- Contrast between faces (famous+unfamiliar) and non-faces (scrambled): difference of averages and significance test.
- Contrast between famous faces and unfamiliar faces: difference of averages and significance test.
- Sensors of interest: EEG070 (or EEG060 or EEG065 if EEG070 is marked as bad)
Expected effects:
- OFA: Bilateral occipital face area
- FFA: Bilateral fusiform face area
- STS: Right superior temporal sulcus
- Around 170ms (N170): Difference between Faces and Scrambled.
- After 250ms: Difference between Famous and Unfamiliar.
The methodology that we will follow for computing the averages and the other statistics is described in the tutorial "Workflows".
Subject averages: Famous, Unfamiliar, Scrambled
We will start by computing the subject-level averages for all the data types we have: sensor-level recordings, source maps and time-frequency results. We will use a weighted average to group the results we have for each run (weighted by the number of good trials). We want to compute the averages for each experimental condition separately (famous, unfamiliar, scrambled).
Since the files are already selected in the interface, we will also compute a grand average across subject. In this context, we will consider that each subject has the same weight in the grand average (option "weighted" not selected). Note that it is not accurate to average MEG recordings across subjects, but we can do it just to get a general idea of the group effects (more information).
MEG/EEG
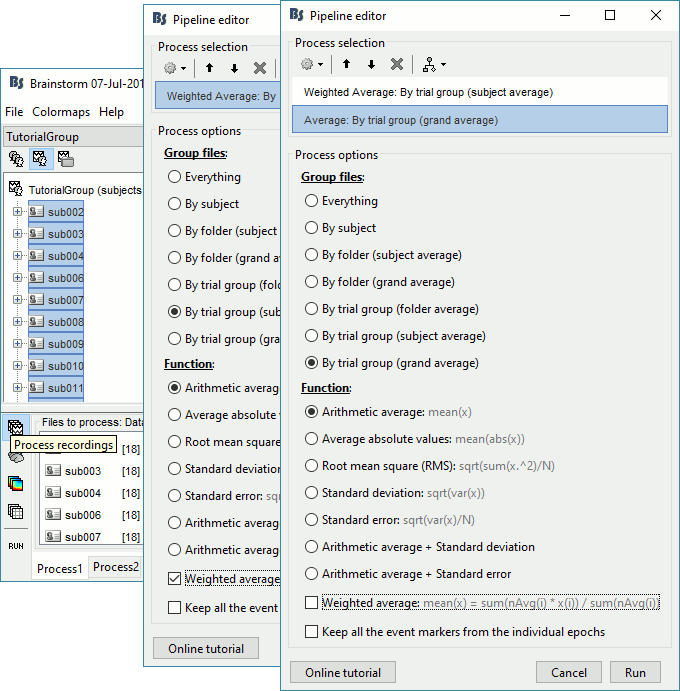
Drag and drop all the subjects in Process1. Select button [Process recordings].
Select process Average > Average files: By trial group (subject average)
Arithmetic average, WeightedAdd process Average > Average files: By trial group (grand average)
Arithmetic average, Not weighted
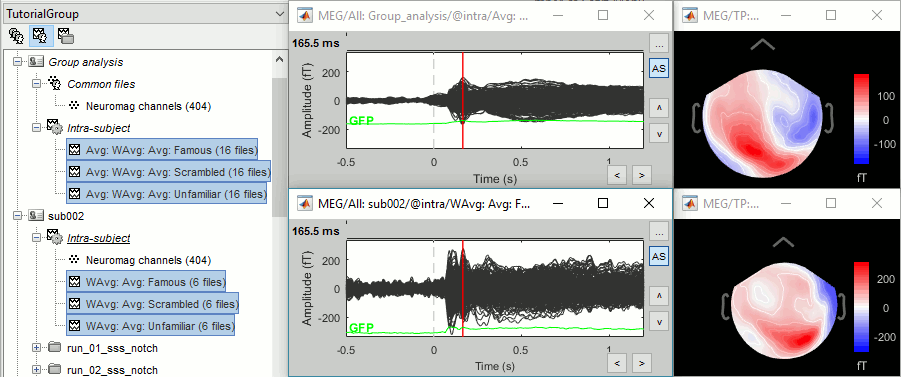
In output, you get three grand averages in the "Group analysis/Inter-subject" folder (top), and three subject averages per subject in the "intra-subject" folders (bottom).
Sources
Keep all the subjects selected in Process1. Select button [Process sources].
If you run the averaging process directly with this file selection, it would average together the MEG and the EEG sources (36 files selected per subject instead of 18). To average separately the two modalities, we can use the Filter box at the bottom-right corner of the Brainstorm window. Enter "EEG" and it would select only the files with a comment including the tag "EEG".
Select process Average > Average files: By trial group (subject average)
EEG source files only, Arithmetic average, WeightedAdd process File > Add tag: EEG.
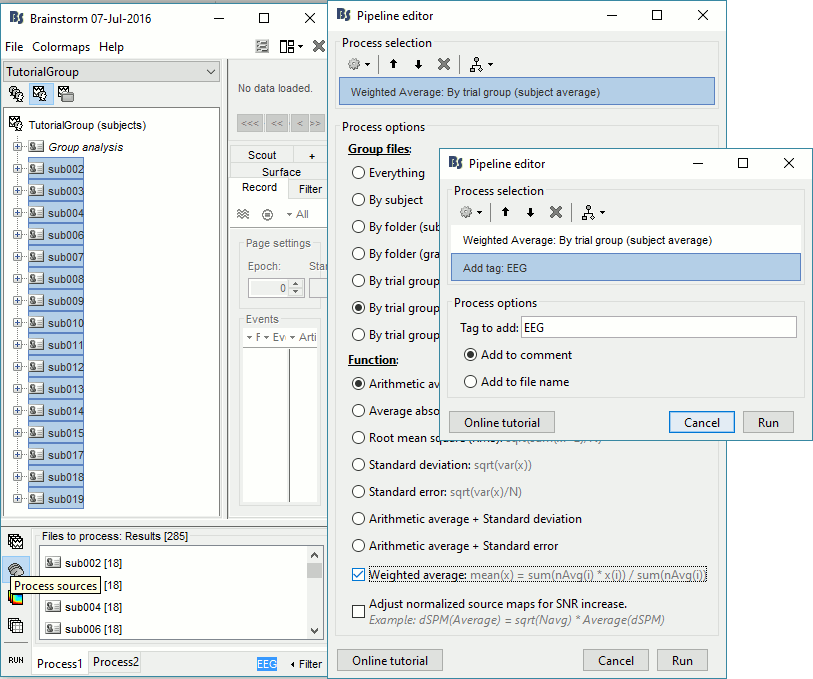
Repeat the same steps with the MEG source files (select all subjects, filter, average, tag).
This time, we cannot compute a grand average directly because the source spaces used to estimate the brain activity do not match between subjects. We have to go through an additional step of projection on a template anatomy. We will also wait to have normalized the source maps before displaying them.
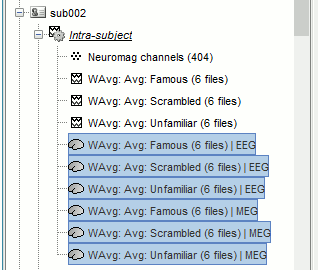
Time-frequency
Keep all the subjects selected in Process1. Select button [Process time-freq].
Make sure you reset the Filter search box at the bottom-right of the Brainstorm window.Run process Average > Average files: By trial group (subject average)
Arithmetic average, Weighted, Match signals
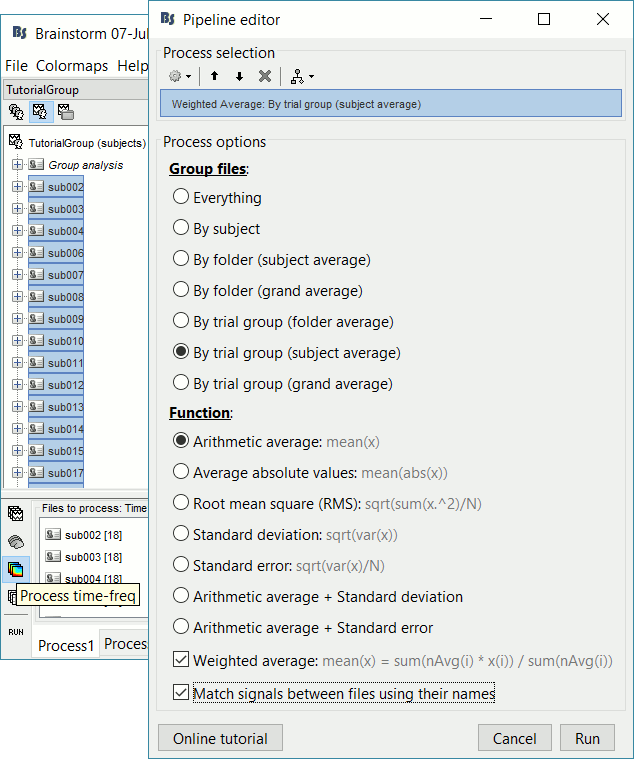
- Like for the sources, we will first normalize the time-frequency maps with respect with a baseline before computing grand averages. Like the sensor data: it is valid to average EEG recordings across subjects, but it is not accurate to average MEG sensors across subjects.
We can average the time-frequency files across runs (it is as valid as averaging the MEG or EEG signals), but we need to select this option "match signals between files using their names" to account for the variable number of bad channels we may have across runs or subjects.
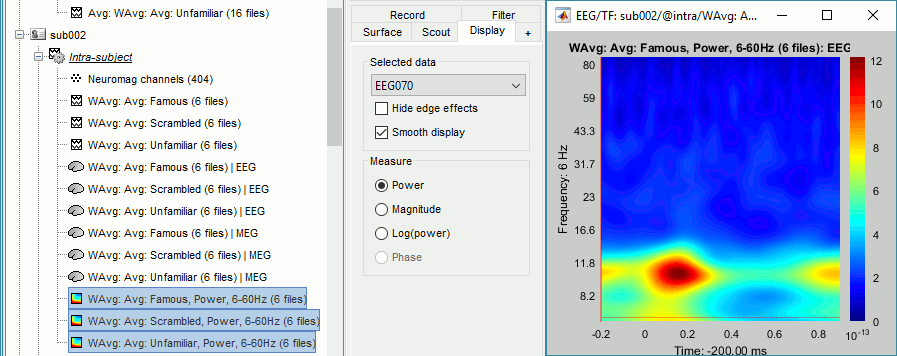
Subject averages: Faces
One of the contrast we want to study is faces (Famous and Unfamiliar) vs non-faces (Scrambled). We need to re-average the Famous and Unfamiliar averages together.
MEG/EEG
- In Process2: FilesA = all the Famous subject averages, FilesB = all the Unfamiliar subject averages. The subjects must be in the same order in the two lists because the process will process the files by pairs of files (A,B).
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces"
Add process Average > Average files: By trial group (subject average): Not weighted
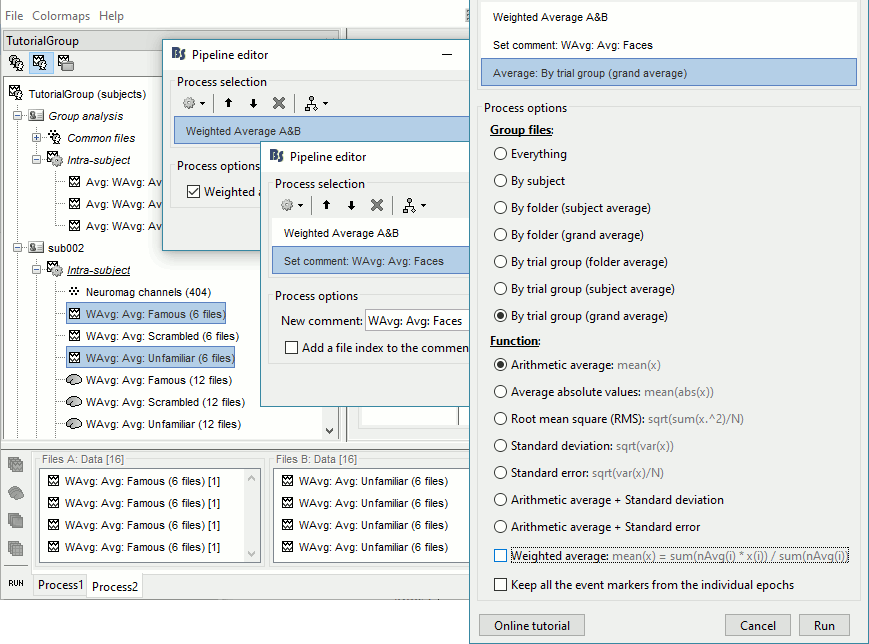
- Selecting that many files from the database explorer can be long when done manually, but can be very efficient from a script. Check out the script equivalent at the end of this page.
It creates one new average for each pair (A,B) and computes the grand average for condition Faces.
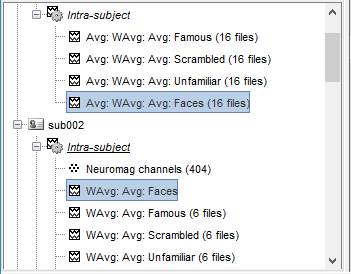
Sources
In Process2: FilesA = all the Famous subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process sources]
- Make sure the order of the files matches in the two lists.
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces | EEG"
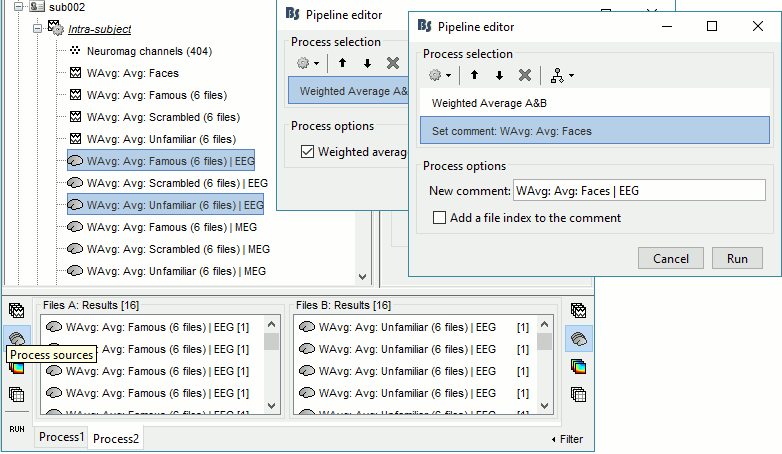
Repeat the same steps for the MEG sources.
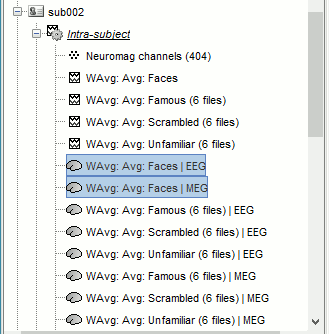
Time-frequency
In Process2: FilesA = all the Famous subject averages (EEG only) [Process timefreq]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process timefreq]
Select process Other > Average A&B: Weighted.
Add process File > Set comment: Comment = "WAvg: Avg: Faces"
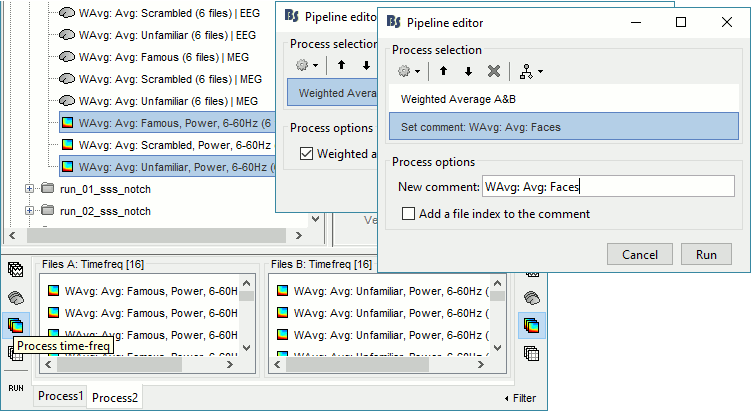
In output: the average for the Face condition, for each subject.

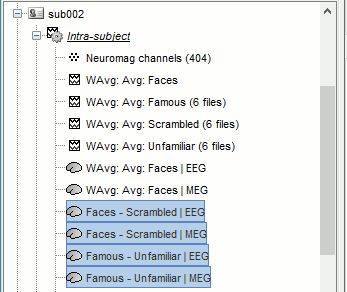
Subject averages: Within-subject differences
To detect correctly the differences between two conditions at the source level, we need to estimate the differences of the conditions for each subject, and then normalize the difference (see tutorial Workflows).
Sources: Faces - Scrambled
In Process2: FilesA = all the Faces subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Scrambled subject averages (EEG only) [Process sources]
Select process Difference > Difference A-B: Do not use absolute values.
Add process File > Set comment: Comment=Faces - Scrambled | EEG
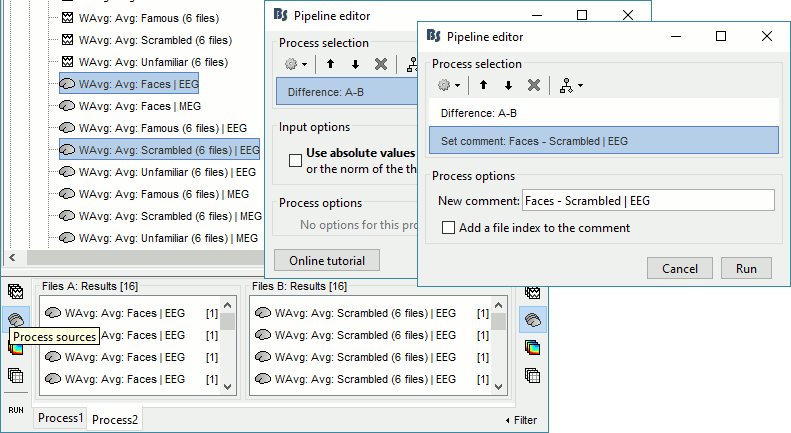
Repeat the same steps for the MEG sources.
Sources: Famous - Unfamiliar
In Process2: FilesA = all the Famous subject averages (EEG only) [Process sources]
In Process2: FilesB = all the Unfamiliar subject averages (EEG only) [Process sources]
Select process Difference > Difference A-B: Do not use absolute values.
Add process File > Set comment: Comment=Famous - Unfamiliar | EEG
Repeat the same steps for the MEG sources.
Subject averages: Filter and normalize
Before comparing the averages across subjects we are going to low-pass filter the signals below 32Hz (to smooth possible latency differences between subjects) and normalize the source and time-frequency values with respect with a baseline (see tutorial Workflows).
MEG/EEG
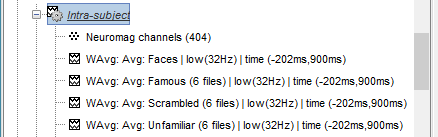
In Process1, select all the Intra-subject folders from all the subjects, select [Process recordings]. For a faster selection, you can use the view "Functional data (sorted by conditions)".
Select process Pre-process > Band-pass filter: 0Hz-32Hz, MEG,EEG, 60dB, No mirror, Overwrite.
Add process Extract > Extract time: Time window=[-200,900]ms
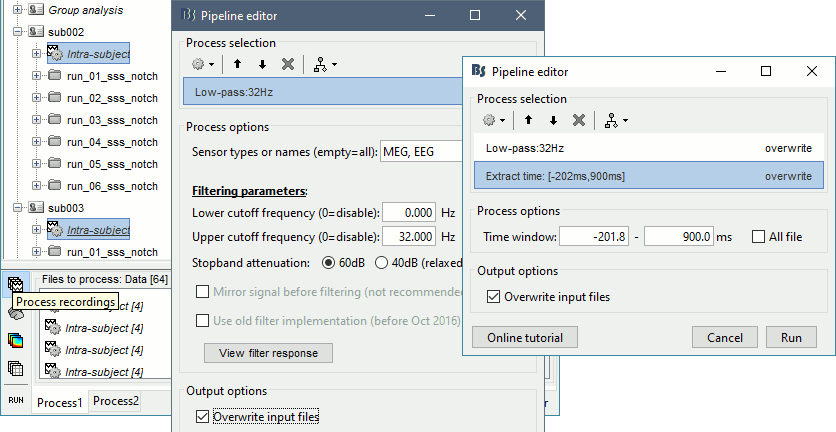
Two tags are added at the end of the comments of the averaged recordings. The selection of a smaller time window gets rid of most of the possible edge effects caused by the filter.
Sources
In Process1, select all the Intra-subject folders from all the subjects, select [Process sources].
Select process Pre-process > Band-pass filter: 0Hz-32Hz, MEG,EEG, Mirror, Overwrite
Add process Extract > Extract time: Time window=[-200,900]ms
Add process Standardize > Baseline normalization: Baseline=[-200,-5]ms, Z-score, Overwrite
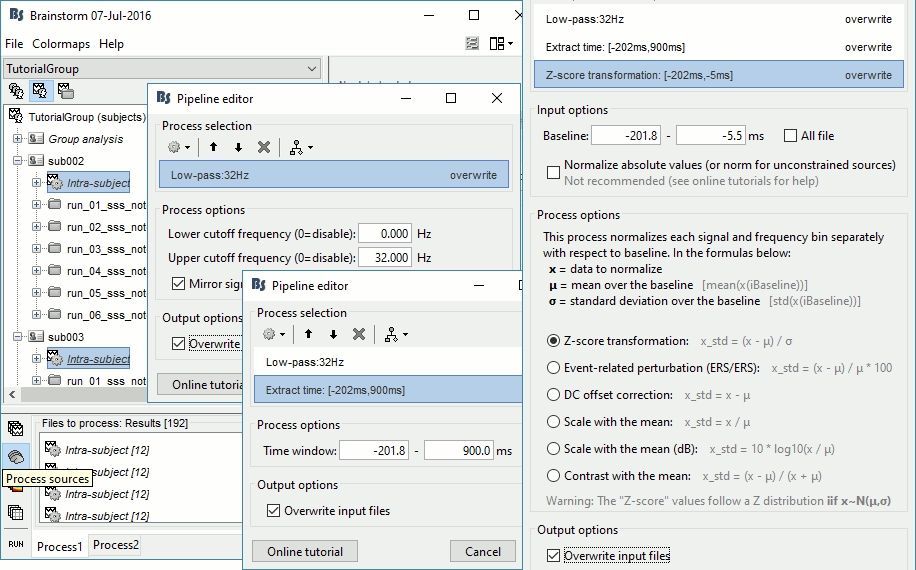
Three tags are added at the end of the comments of the averaged sources.

Time-frequency
In Process1, select all the Intra-subject folders from all the subjects, select [Process time-freq].
Run process Standardize > Baseline normalization: Baseline=[-200,-5]ms, ERS/ERD, Overwrite
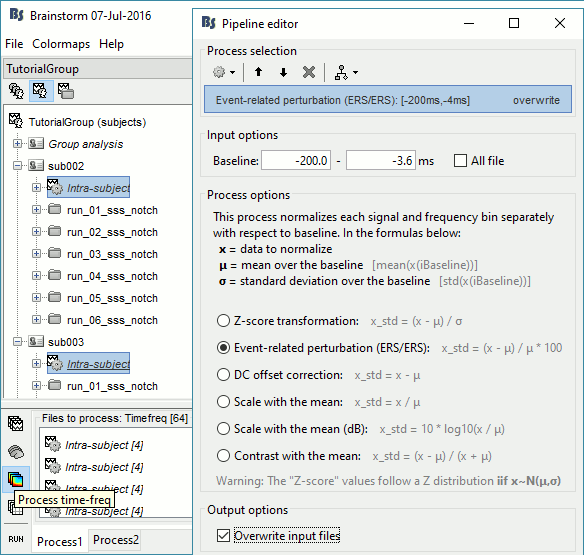
One tag is added at the end of the comments of the averaged time-frequency maps.

Subject averages: Screen captures
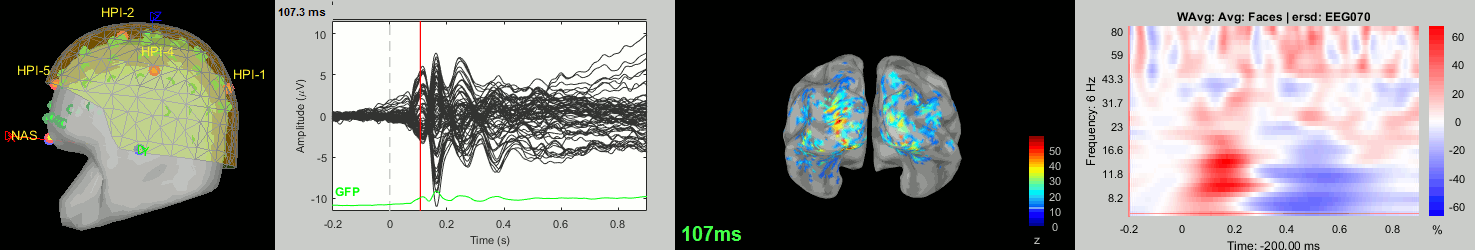
Now we have all the measures ready to be compared across subjects: MEG/EEG, sources, time-frequency. Let's take a few screen captures to make sure the primary visual response look good for all the subjects. The screen captures below represent from left to right:
- Registration sensors/MRI: Head surface, MEG (inside of the helmet), EEG+headpoints (green points).
- Faces: EEG recordings
- Faces: MEG sources at 107ms
- Faces: Time-frequency for EEG070
sub002
sub003
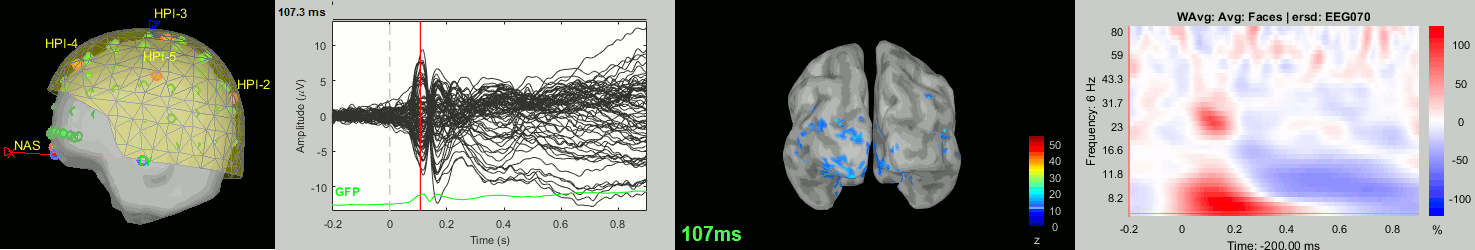
sub004
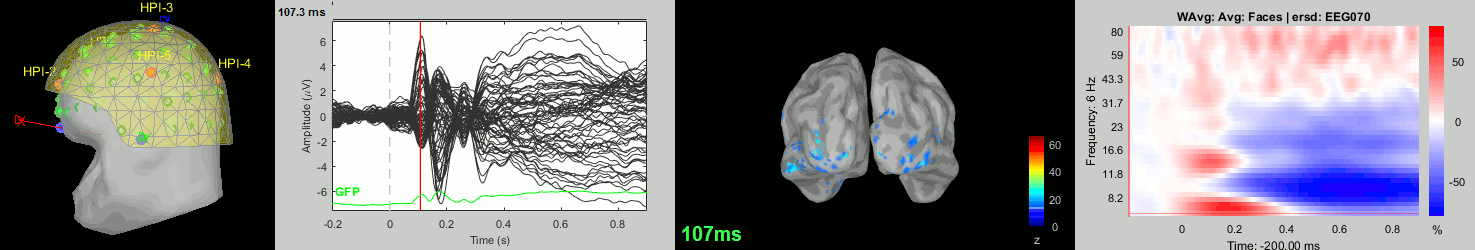
sub006
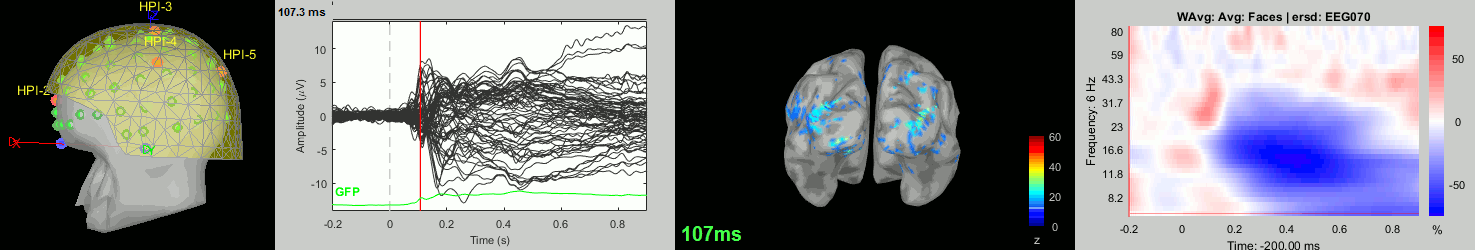
sub007
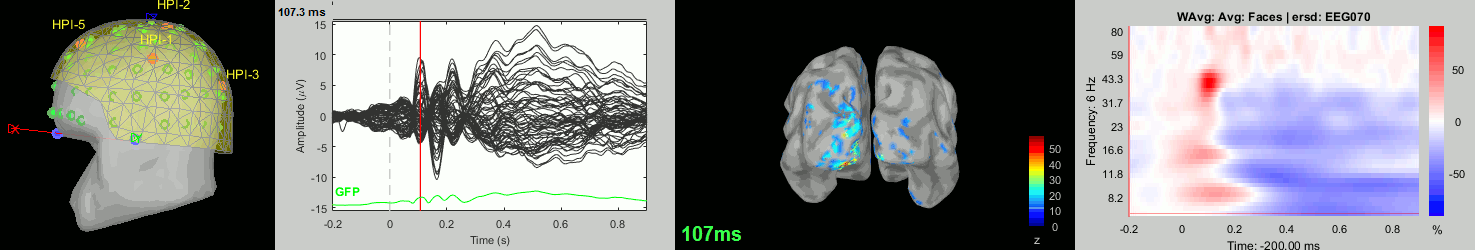
sub008
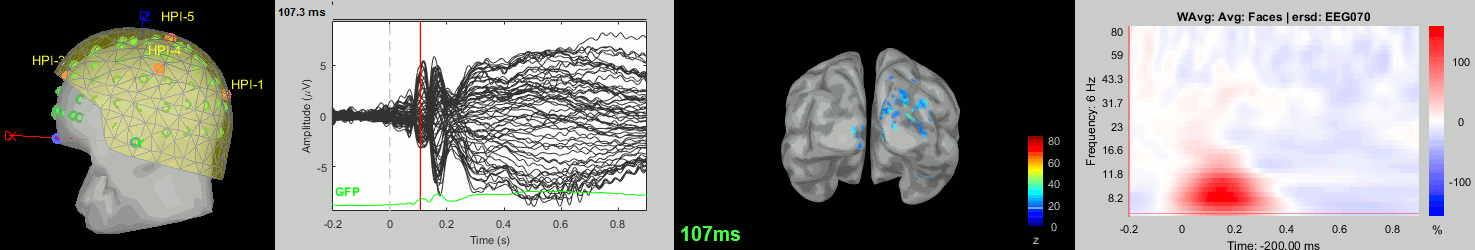
sub009
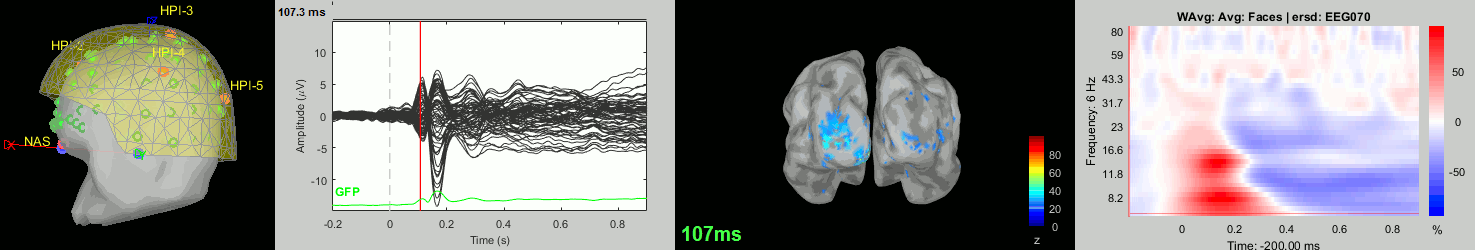
sub010
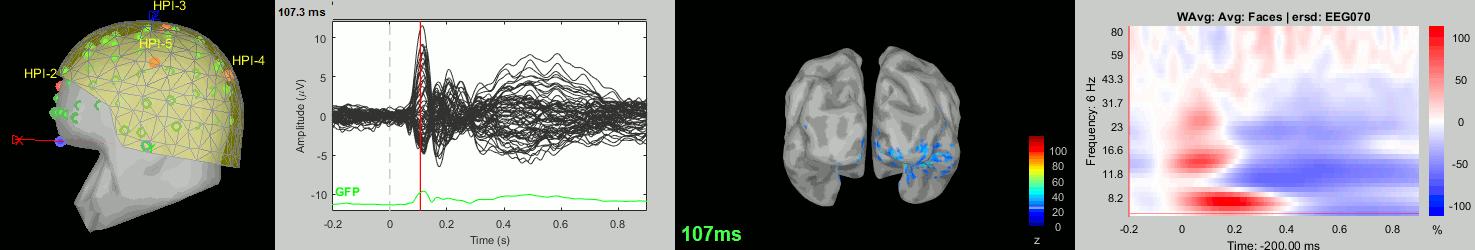
sub011
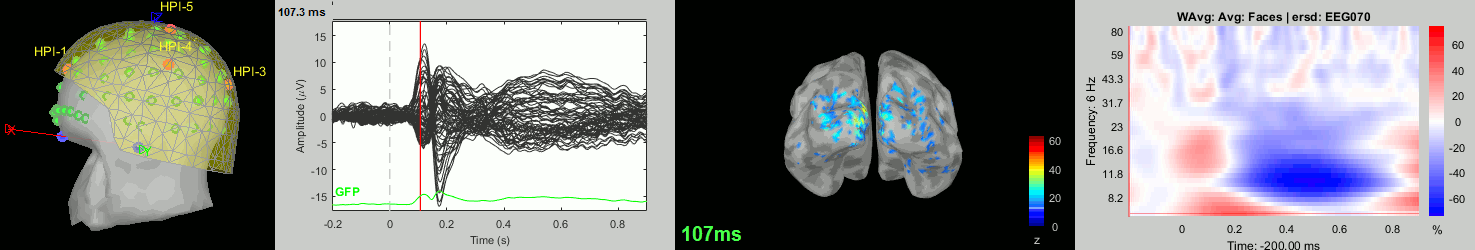
sub012
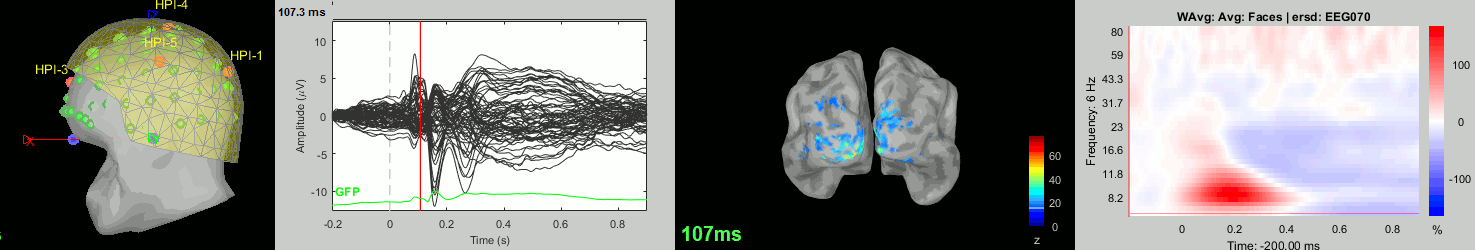
sub013
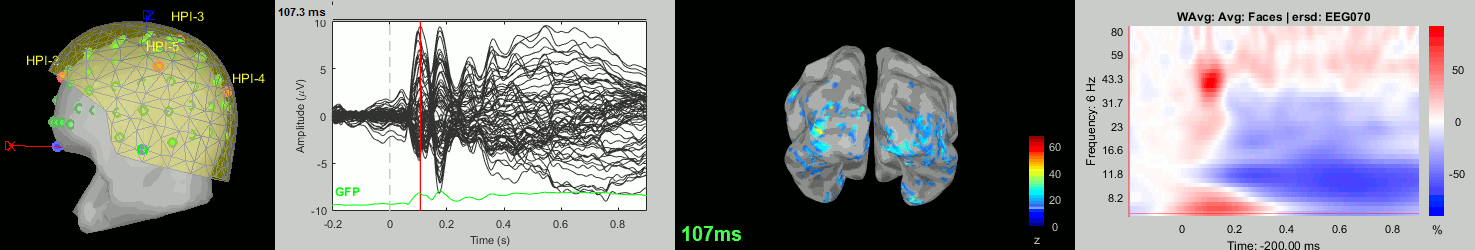
sub014
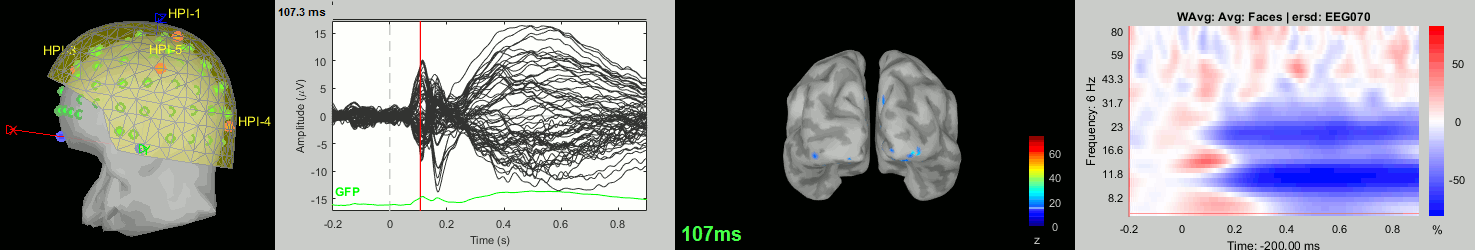
sub015
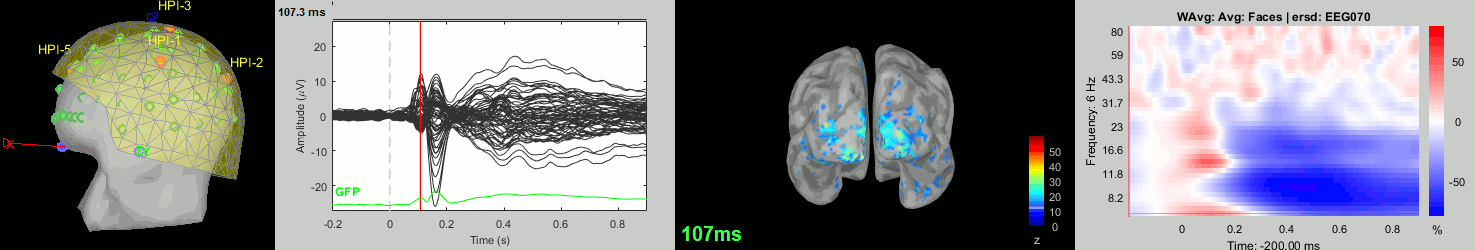
sub017
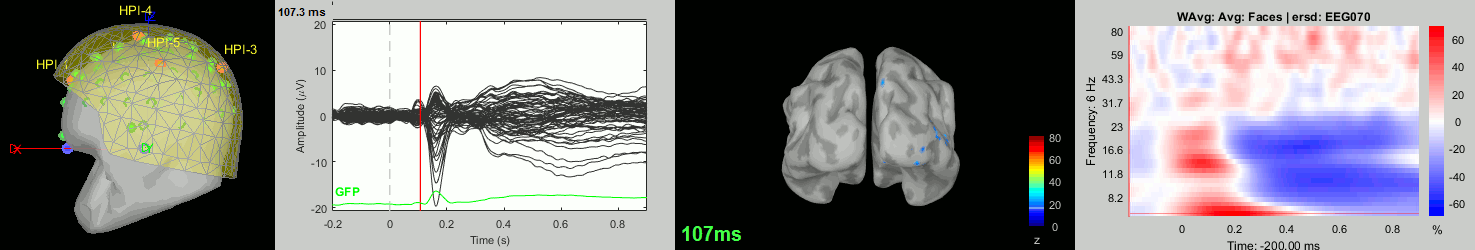
sub018
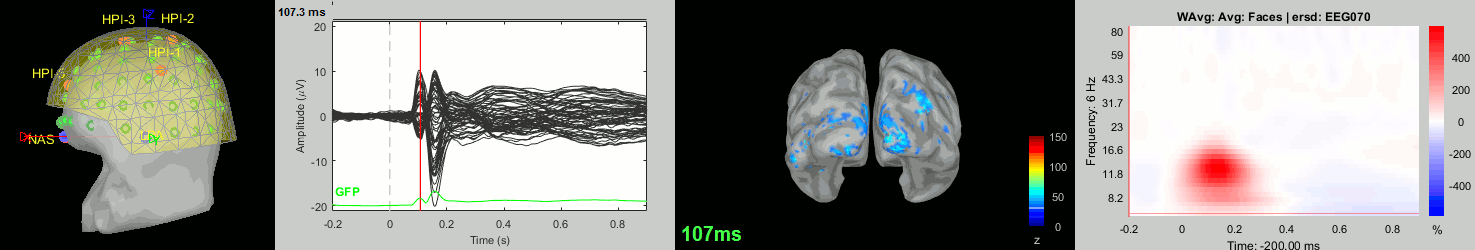
sub019
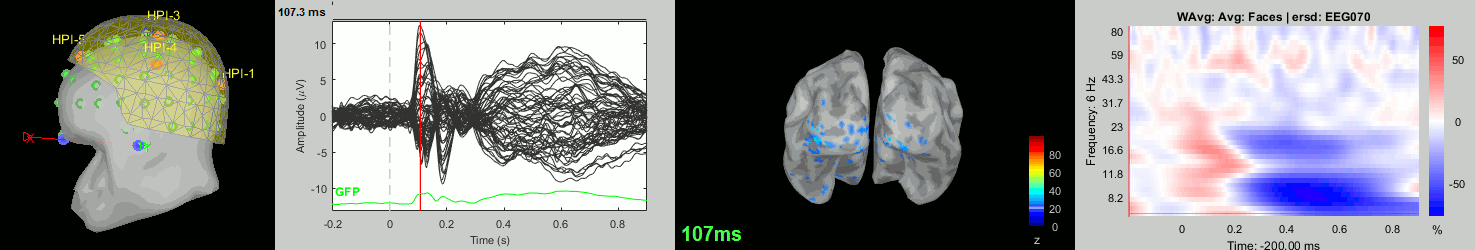
Group analysis: MEG/EEG
Grand averages
We have already computed the group averages for all the conditions. We will not look much further at these results as we are more interested in the contrasts between conditions. Below are screen captures for all group averages (top=MEG MAG, bottom=EEG). Topography at [50,100,150,200,250,300]ms.
Faces: Famous / Unfamiliar
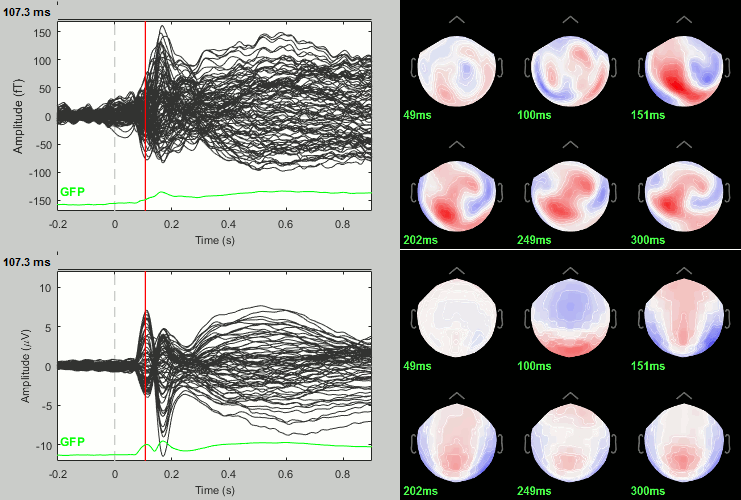
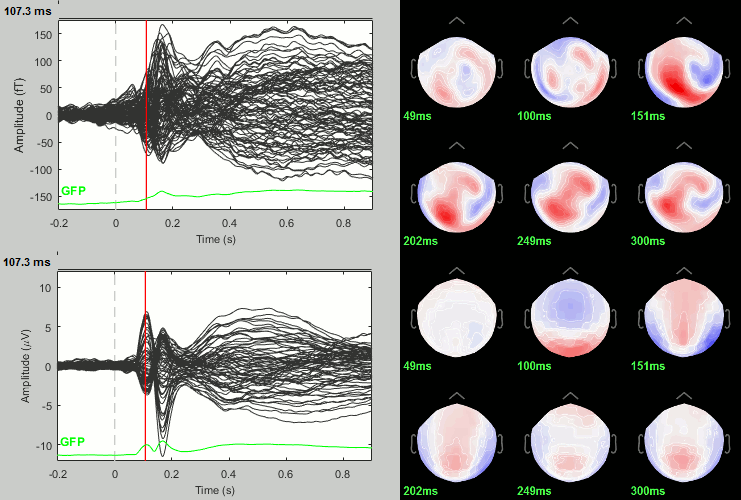
Scrambled
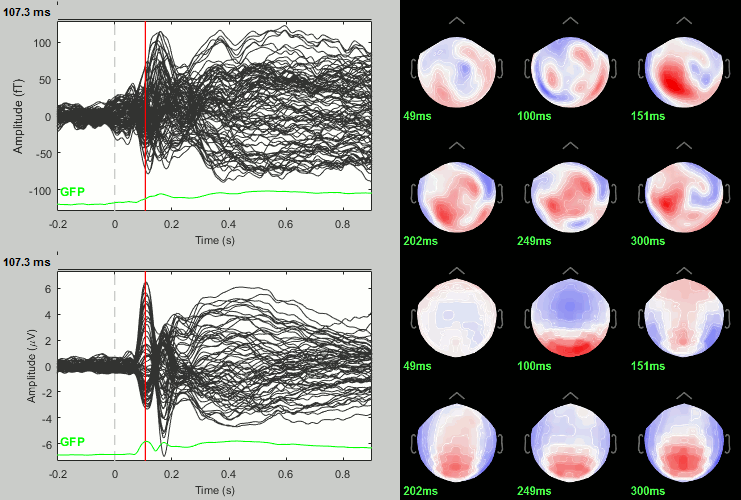
Faces - Scrambled: Differences of averages
We could compute the contrasts directly from the grand averages, but we will do it from the subject averages because it will be the same file selection for the statistics.
- In Process2: FilesA = all the Faces subject averages (from the Intra-subject folders).
- In Process2: FilesB = all the Scrambled subject averages (from the Intra-subject folders).
Run process: Test > Difference of means: Arithmetic average, Not weighted.
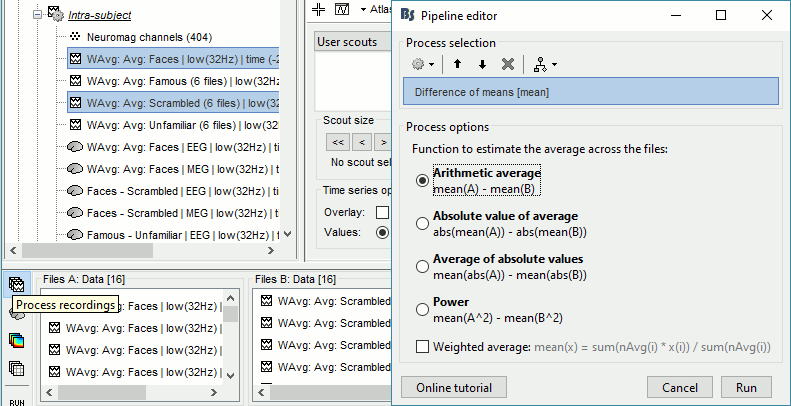
Rename the file: Faces - Scrambled (top=MEG MAG, bottom=EEG).
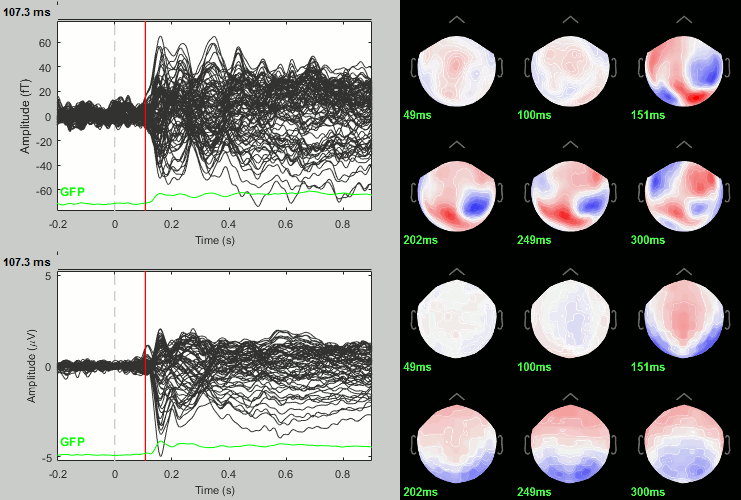
Faces - Scrambled: Significance testing
We have computed the amplitude of the difference between the two conditions, and we will try now to estimate a p-value indicating the significance of each of these values. In all the screen captures below: top=MEG MAG, bottom=EEG.
- In Process2: Keep the same file selection.
Run process: Test > Parametric test: Paired: All file, All sensors, No average, two-tailed.
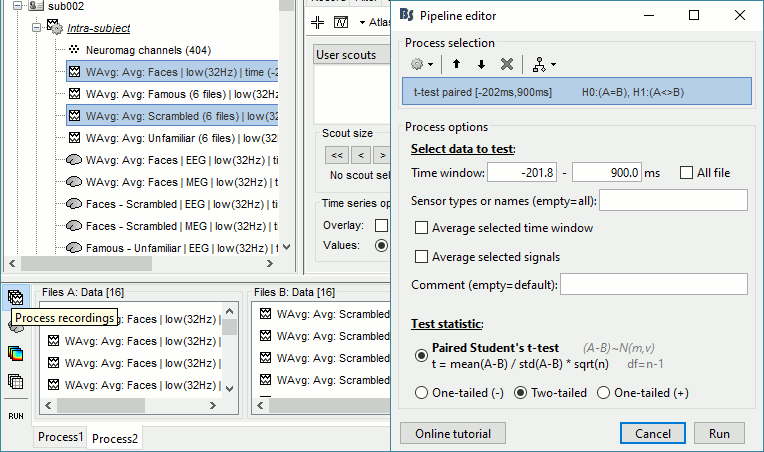
Rename the file: Faces - Scrambled: Parametric t-test. Display with p<0.05, FDR-corrected.
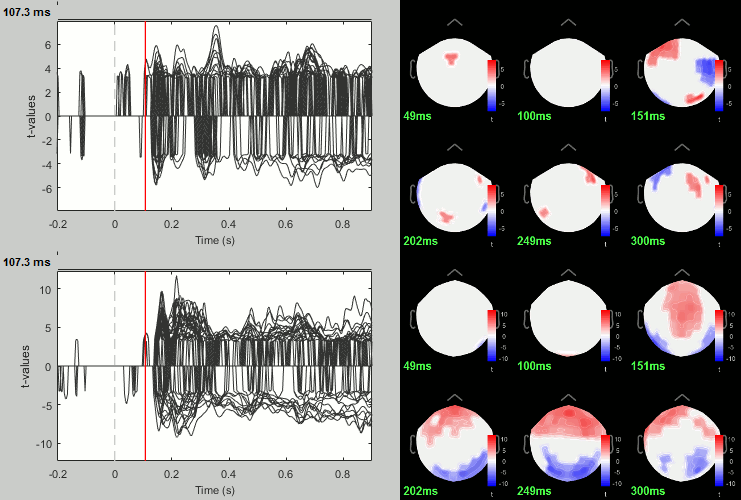
- You can run other tests in a similar way, with almost identical results.
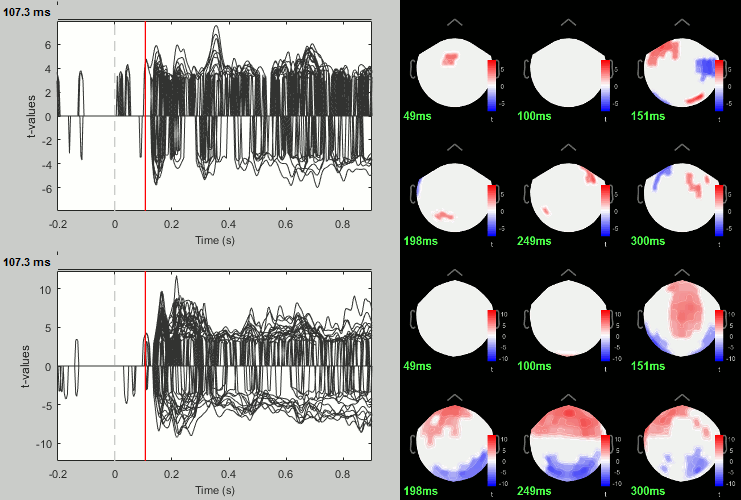
Process: Test > Permutation test: Paired: All file, All sensors, Paired t-test, 1000 randomizations. Display with p<0.05, FDR-corrected.
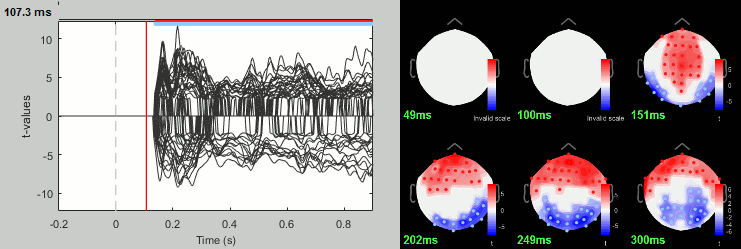
Process: Test > FieldTrip: ft_timelockstatistics: All file, EEG, Paired t-test, 1000 randomizations, correction=cluster, cluster alpha=0.05. The cluster-based statistics must be executed on one type of sensors at a time (EEG, MEG MAG or MEG GRAD), because it tries to identify spatio-temporal clusters which groups adjacent sensors.
Famous - Unfamiliar: Differences of averages
- In Process2: FilesA = all the Famous subject averages (from the Intra-subject folders).
- In Process2: FilesB = all the Unfamiliar subject averages (from the Intra-subject folders).
Run process: Test > Difference of means: Arithmetic average, Not weighted.
Rename the file: Famous - Unfamiliar.
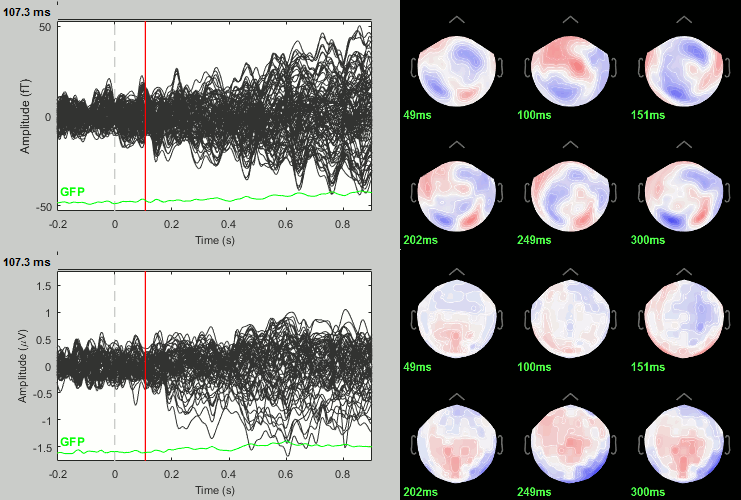
Famous - Unfamiliar: Significance testing
- In Process2: Keep the same file selection.
Run process: Test > Parametric test: Paired: All file, All sensors, No average, two-tailed.
Rename the file: Faces - Scrambled: Parametric t-test. Display with p<0.05, FDR-corrected.
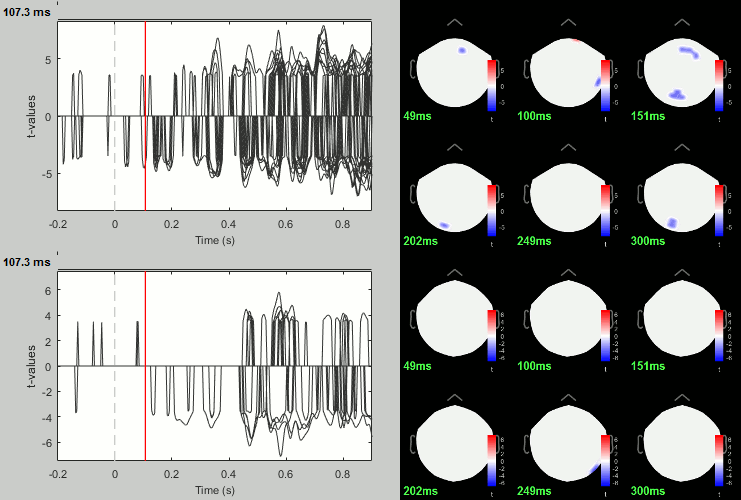
Run process: Test > FieldTrip: ft_timelockstatistics: All file, EEG, Paired t-test, 1000 randomizations, correction=cluster, cluster alpha=0.05.
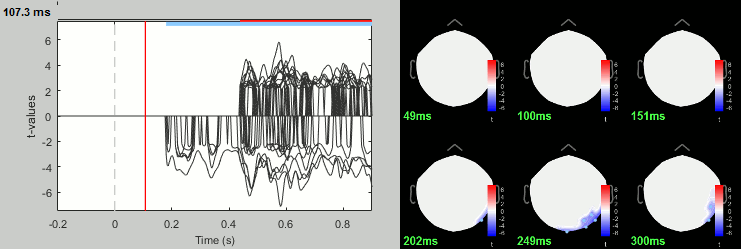
Group analysis: Sources
Projet sources on template
The sources were estimated on the individual anatomy of each subject, the resulting cortical source maps cannot be averaged directly. We need first to re-interpolate all the individual results on a common template (the ICBM152 brain, available in the "default anatomy" folder of the protocol). We also need to extract the absolute values for these source maps: the sign of the minimum norm maps are relative to the orientation of the current with respect to the surface normal, which can vary between subjects.
In Process1, select all the Intra-subject folders from all the subjects, select [Process sources]. For a faster selection, you can use the view "Functional data (sorted by conditions)".
Select process Pre-process > Absolute values: Sensor types=MEG,EEG, Overwrite.
Add process Sources > Project on default anatomy.
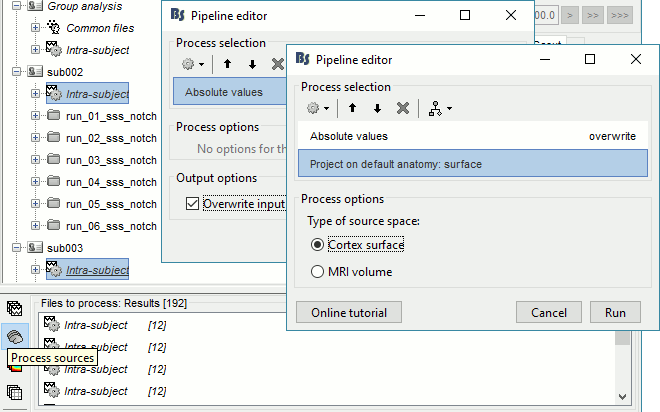
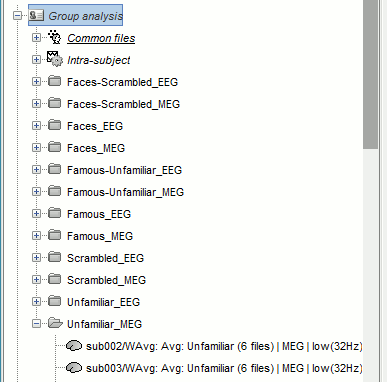
- All the 192 subject averages are projected in the same folder "Group analysis / Intra-subject". Re-organize all the projected source files in new folders, one folder per condition.
To create a folder, right-click on Group analysis > New folder. To move a file to the new folder: drag and drop it in the database explorer or use the keyboard shortcuts Ctrl+X/Ctrl+V.
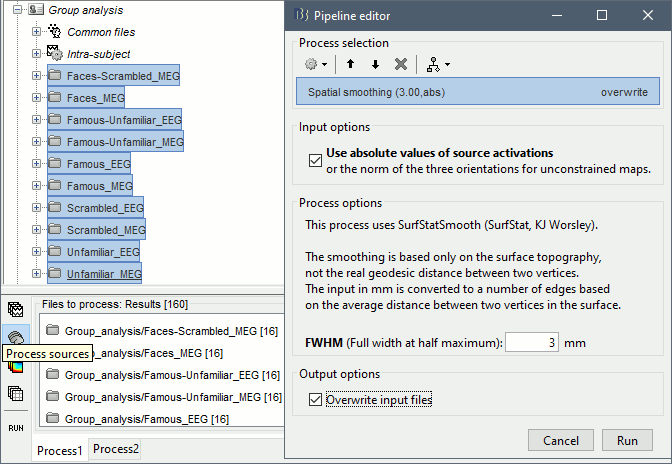
Spatial smoothing
The source maps estimated with constrained orientations can show very focal activity: two adjacent vertices may have very different normals, and therefore very different current values. When averaging multiple subjects, the peaks of activity may not align very well across subjects. Smoothing spatially the source maps may help obtaining better group results.
- In Process1, select all the source maps in Group analysis.
Run process Sources > Spatial smoothing:
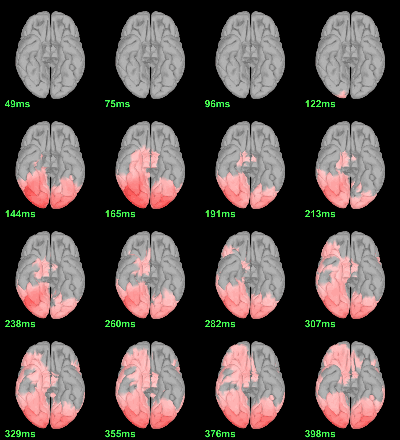
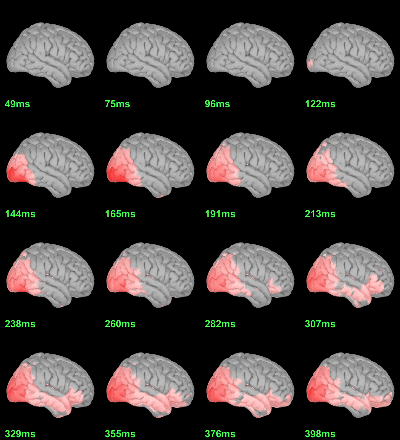
MEG: mean(|Faces-Scrambled|)
- In Process1, select all the source files in Group_analysis/Faces-Scrambled_MEG.
Run process Average > Average files: By folder (grand average), Not weighted.
Display with an arbitrary threshold: Z-score>3.
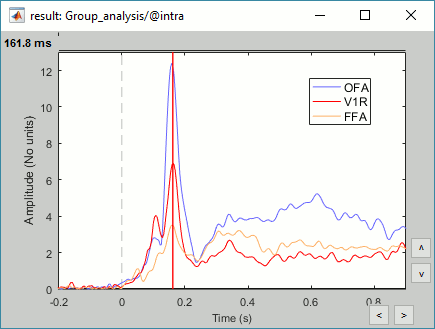
Regions of interest: OFA (Occipital Face Area), FFA (Fusiform Face Area), V1.
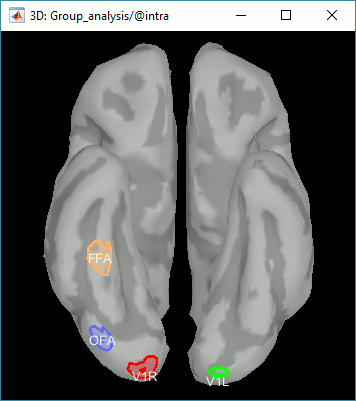
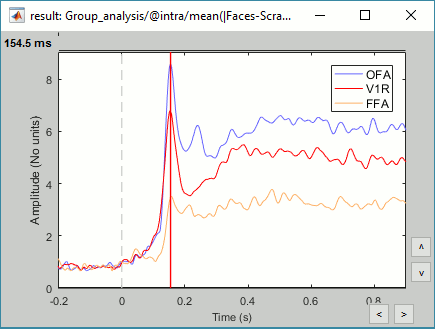
- This approach identifies correctly the effect size at the subject level. But because the sign is ambiguous across subjects, we need to average the absolute values of the differences. Therefore we are able to identify when and where the two conditions are different, but not which condition leads to a stronger brain response.
MEG: Chi2-test |Faces-Scrambled|=0
- In Process1, select all the source files in Group_analysis/Faces-Scrambled_MEG.
Run process Test > Parametric test against zero: All file, One-sample Chi2-test two-tailed.
Screen capture: p<0.05 FDR-corrected
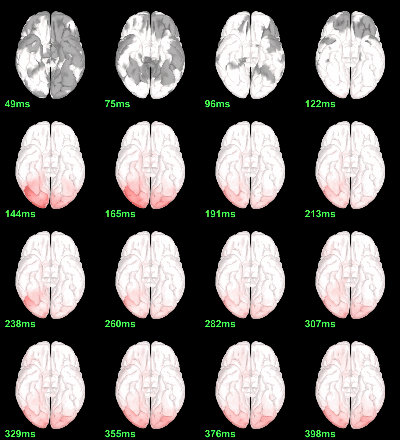
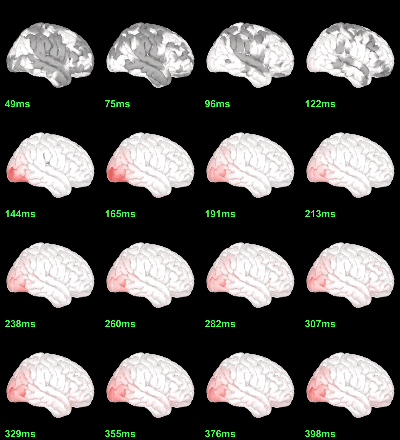
This tests the hypotesis H0:[mean_subject(|Faces-Scrambled|) = 0]
This test is appropriate to detect the differences between the two conditions, but cannot determine which condition is stronger. Its obvious limitation is that it is too sensitive: post-stimulation, everything is shown as significant.
MEG: Chi2-test log(|Faces-Scrambled|)=0
Right-click on the folder Group_analysis/Faces-Scrambled_MEG > File > Duplicate folder.
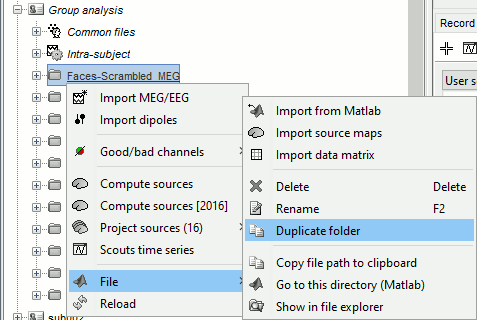
Rename the new folder: Faces-Scrambled_MEG_log.
- In Process1, select all the source files in Group_analysis/Faces-Scrambled_MEG_log.
Select process Pre-process > Run Matlab command > "Data = log(Data)";
Add process Test > Parametric test against zero: All file, One-sample Chi2-test two-tailed.
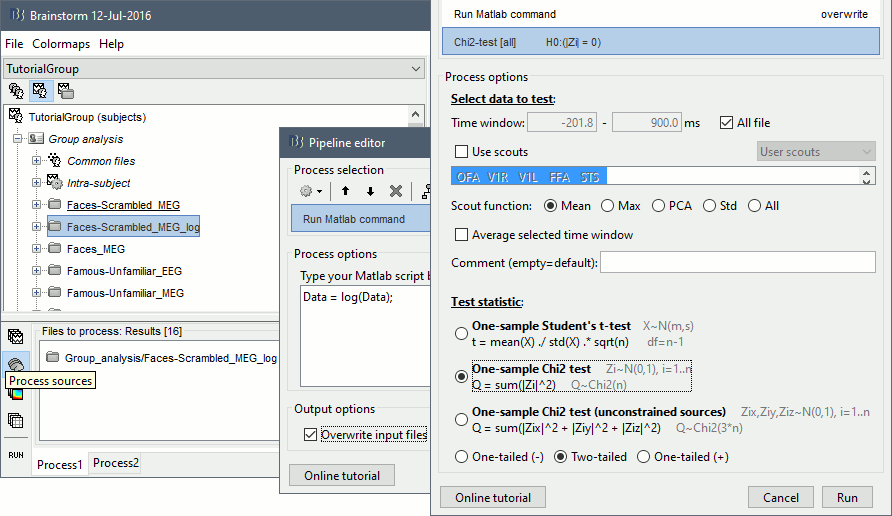
Screen capture: p<0.05, FDR-corrected
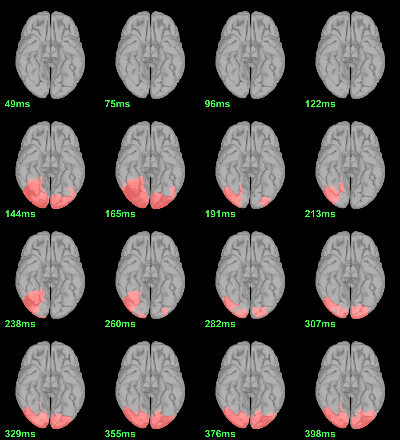
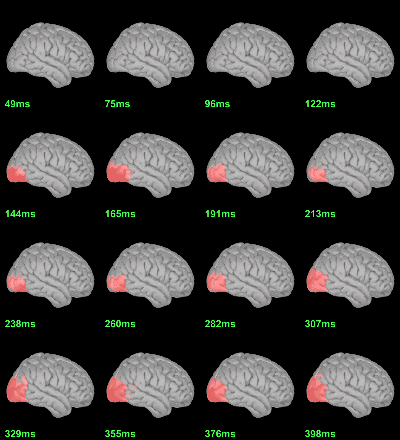
MEG: mean(|Faces|)-mean(|Scrambled|)
- In Process2: FilesA = all the source files in Group_analysis/Faces_MEG.
- In Process2: FilesB = all the source files in Group_analysis/Scrambled_MEG.
Run process Test > Difference of means: Arithmetic average, Not weighted.
Display with an arbitrary threshold: Z-score>3
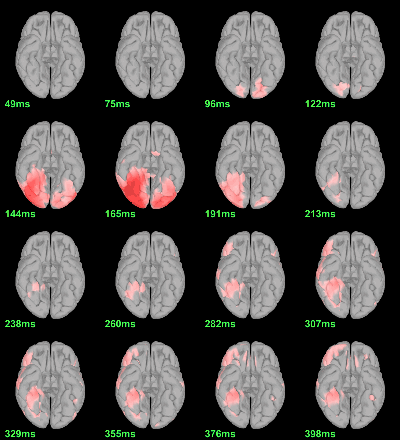
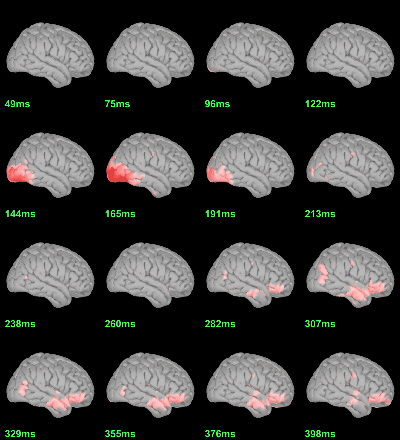
Regions of interest: OFA (Occipital Face Area), FFA (Fusiform Face Area), V1.

- This approach is not able to detect correctly all the differences between the two conditions. At the subject level,we discarded the sign of each condition, therefore we are not able to detect differences in the orientation of the sources or phase delays between conditions.
MEG: Student t-test |Faces|=|Scrambled|
- In Process2: FilesA = all the source files in Group_analysis/Faces_MEG.
- In Process2: FilesB = all the source files in Group_analysis/Scrambled_MEG.
Run process Test > Parametric test: Paired: All file, No average, two-tailed.
Screen capture: p<0.05 FDR-corrected
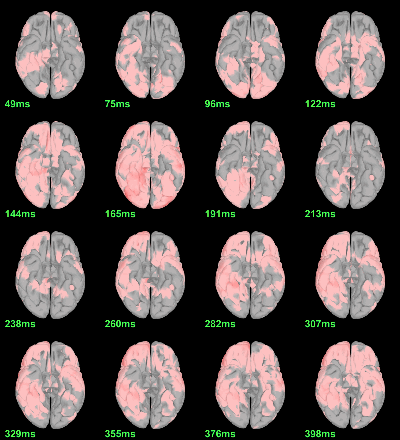
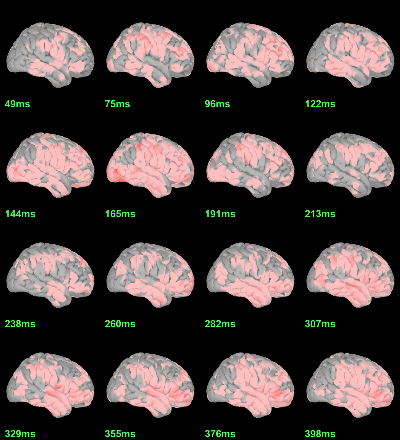
This test the hypothesis H0:[mean(abs(Faces))=mean(abs(Scrambled))].
This measure is not appropriate to properly identify the differences between the two conditions because it is insensitive to the sign of the sources within each subject.
EEG: Faces-Scrambled
- Repeat the same operations with the EEG sources.
Difference of averages: |Faces-Scrambled| (Z-score>5):
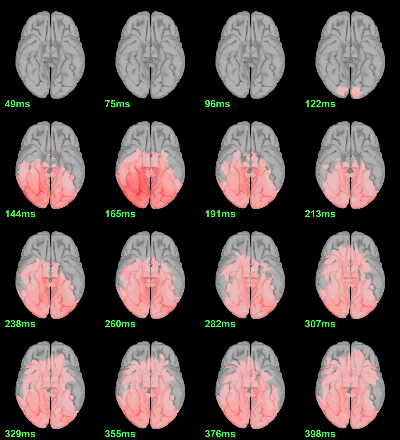
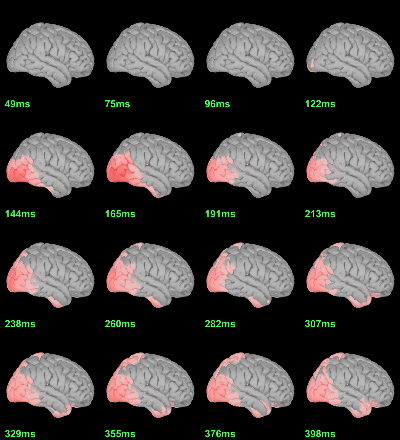
Parametric Chi2-test: mean_subject(|Faces-Scrambled|) = 0, p<0.05 FDR-corrected
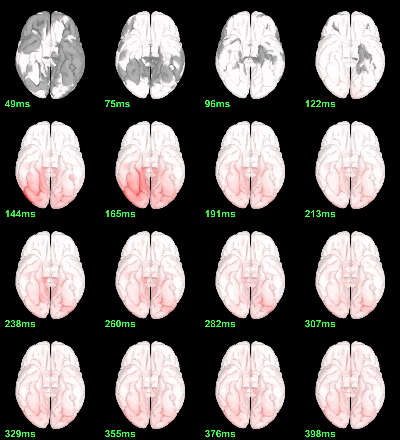
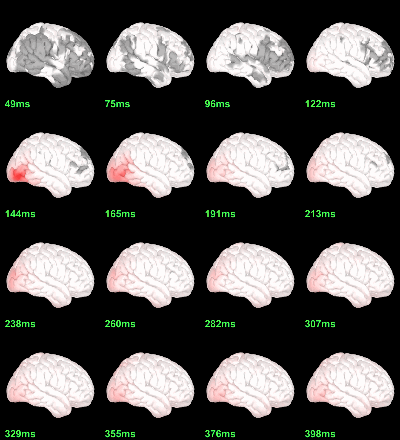
Parametric Chi2-test: log(mean_subject(|Faces-Scrambled|)) = 0, p<0.05 FDR-corrected
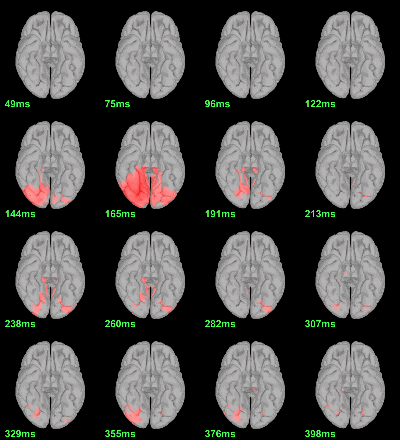
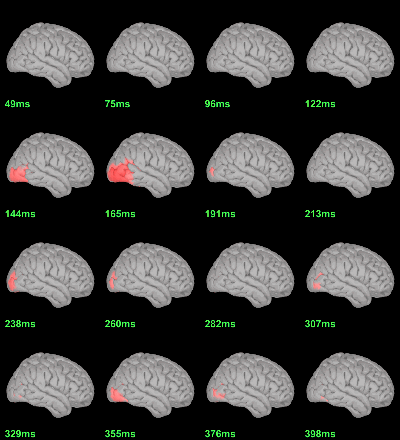
Difference of averages: |Faces|-|Scrambled| (Z-score>3)
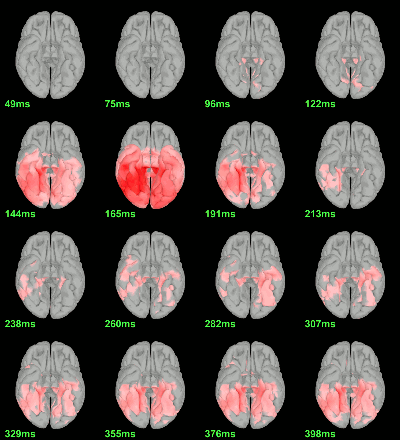
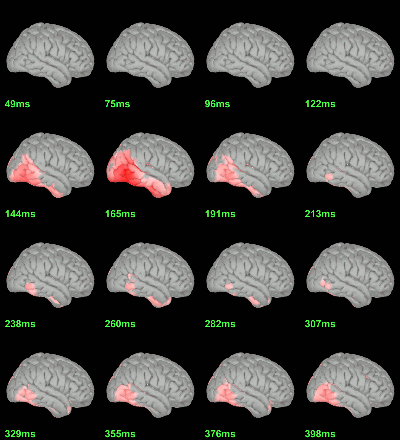
Parametric Student t-test: mean(abs(Faces))=mean(abs(Scrambled)), p<0.05 FDR-corrected
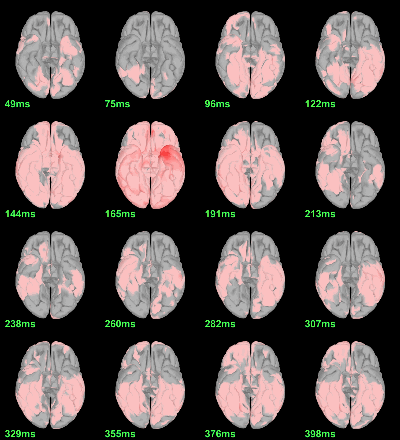
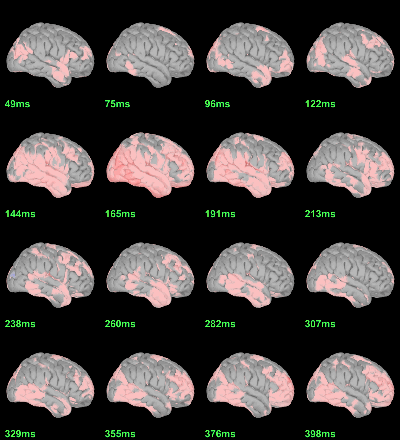
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_visual_group.m