|
Size: 15712
Comment:
|
Size: 15720
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| ''Authors: Francois Tadel, Ei-ichi Okumura, Takashi Asakawa, Yasuhiro Haruta.'' | ''Authors: Francois Tadel'''''', Yasuhiro Haruta'', Ei-ichi Okumura, Takashi Asakawa.'' |
Yokogawa/KIT tutorial
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Yasuhiro Haruta, Ei-ichi Okumura, Takashi Asakawa. This tutorial introduces some concepts that are specific to the management of MEG/EEG files recorded with Yokogawa/KIT systems in the Brainstorm environment. Contents
This tutorial dataset (MEG/EEG and MRI data) remains proprietary of Yokogawa Electric Corporation, Japan. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from Yokogawa Electric Corporation.
This tutorial is based on a simple median nerve stimulation experiment:
To import Yokogawa/KIT data files (.con, .raw, .ave) into Brainstorm, a data export process is required beforehand. The data export function is available in Meg160, which is data analysis software equipped in most of Yokogawa/KIT systems. The dataset used in this tutorial has already been exported using this procedure. It is described here so that later you can export your own recordings to Brainstorm. If your software does not support the functions used below, please contact Yokogawa via Export the digitizer file In Meg160, select the menu: File > Import and Export > BESA Text Export > Surface Point File Export the recordings In Meg160, select the menu: File > Import and Export > Third-Party Export Enter the digitizer file in the [Point Filename] box Enter the label file in the [Label Filename] box
Go to the Download page of this website, and download the file: sample_yokogawa.zip Unzip it in a folder that is not in any of the Brainstorm folders (program or database folder) Select the menu File > Create new protocol. Name it "TutorialYokogawa" and select the options: "No, use individual anatomy", "No, use one channel file per condition".
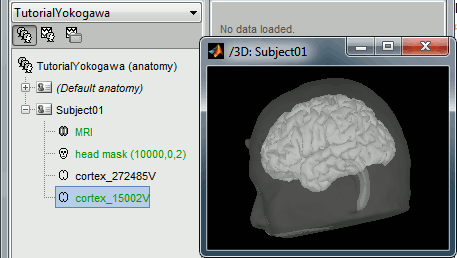
Right-click on the TutorialYokogawa folder > New subject > Subject01 Right-click on the subject node > Import anatomy folder: Set the file format: "FreeSurfer folder" Select the folder: sample_yokogawa/anatomy At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, which will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.
If you do not have access to an individual MR scan of the subject (or if its quality is too low to be processed with FreeSurfer), but if you have digitized the head shape of the subject using a tracking system, you have an alternative: deform one of the Brainstorm templates (Colin27 or ICBM152) to match the shape of the subject's head.
Right-click on the subject folder > Review raw file: Select the file: sample_yokogawa/data/SEF_000-export.con Answer NO when asked to refine the registration using head points. In this dataset, we only have access to the positions of the electrodes and three additional markers on the forehead. The automatic registration doesn't work well in this case, we are going to fix this registration manually.
Right-click on the channel file > Edit channel file: Channel EO1 (208) and EO2 (209): Change the type to EOG Channel EKG+ (214): Change the type to ECG Channel E (231): Change the type to MISC SCREEN CAPTURE
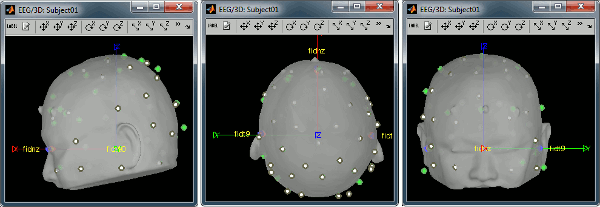
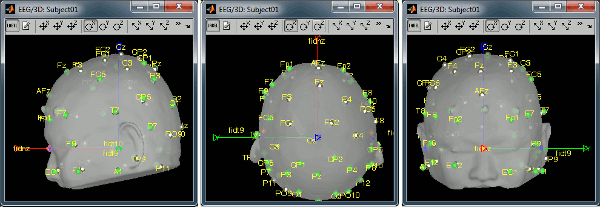
Right-click on the channel file > MRI registration > Edit... (EEG) Now try to manipulate the position of the EEG+MEG sensors using rotations and translations only (no "resize" or individual electrodes adjustments). The objective is to have all the points close to the surface and the three forehead points inside the little peaks on the surface (due to markers in the MRI). The rotation+translation are going to be applied both to the EEG and the MEG sensors. After you are done with this solid registration part, you can click on the button "Project electrodes on scalp surface", it will help for the source modeling later. The green points (digitized) stay in place, the white points (electrodes) are now projected on the skin of the subject. Click on [OK] when done. Answer YES to save the modifications. Answer YES again to apply the solid transformation (rotation+translation) to the MEG sensors. Before manual registration: After manual registration:
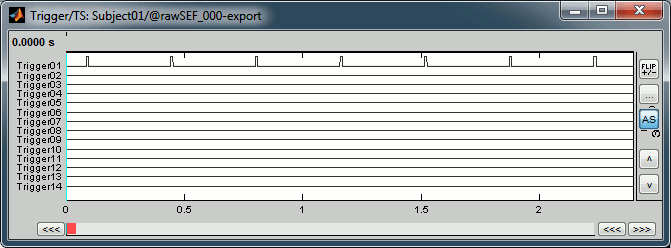
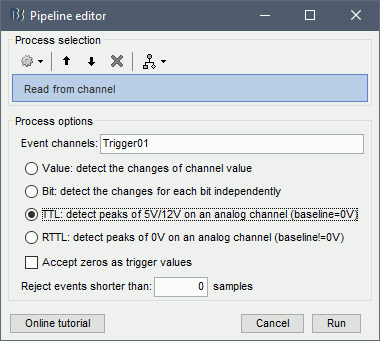
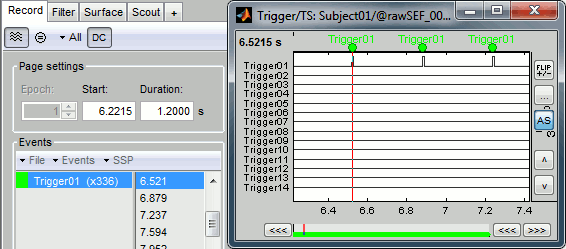
Right-click on the "Link to raw file" > Trigger > Display time series You can see that all the trigger lines are flat except for "Trigger01", which contains the information of the electric stimulation. We are going to read this trigger channel and convert it to a list of events. Close this figure. Run the process "Import recordings > Read from channel" Event channel = Trigger01 Option selected "TTL": detect peaks of 5V/12V on an analog channel Do not select the option Accept zeros as trigger values Right-click on the "Link to raw file" > Trigger > Display time series. Check that the peaks of the triggers channel have correctly been identified, then close this figure.
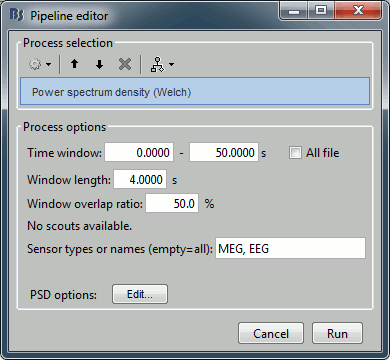
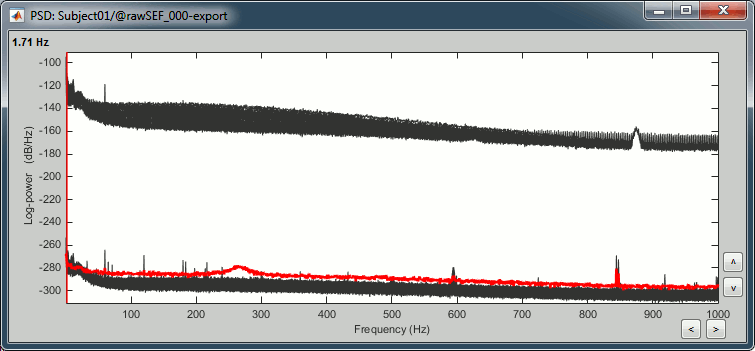
Two of the typical pre-processing steps consist in getting rid of the contamination due to the power lines (50 Hz or 60Hz) and of the frequencies we are not interested in (a low-pass filter to remove the high-frequencies and a high-pass filter to remove the very slow components of the signals). Let's start with the spectral evaluation of this file. Drag the "Link to raw file" to the Process1 box and run the process "Frequency > Power spectrum density (Welch)". Configure it as illustrated in the following figure (window length=10s, overlap=50%). Double-click on the new PSD file to display it. Peaks at 60Hz, 120Hz, 180Hz, 240Hz on EEG + MEG: Peaks at 35Hz, 65Hz, 70Hz, 183Hz, 197Hz on MEG only: MEG sensor LC11 appears to have a higher level of noise than all the other MEG sensors, we will check this when review the MEG recordings and probably tag it as a bad channel.
There is no easy way to process the Yokogawa files at this time using the Brainstorm software. This capability might be added in the future, in which case the procedure would be very similar to the one presented in the basic tutorial ?Detect and remove artifacts. For short recordings, if the contamination is really bad, there is an alternative option: importing the entire file in Brainstorm and then process it. This approach is illustrated in the EEG/Epilepsy tutorial. This approach requires a very large amount of memory, it is not recommended on MEG recordings as the files tend to be much bigger than EEG-only recordings.
Run process "Artifacts > Detect eye blinks" on channel EOG1 and/or EOG2 Run process "Artifacts > Detect heartbeat" on channel EKG+ Run process "Artifacts > SSP: Eye blinks" for MEG and/or EEG Run process "Artifacts > SSP: Heartbeat" for MEG and/or EEG
Right-click on the "Link to raw file" > MEG > Display time series. Click on the noisy LC11 sensor to select it (displayed in red) Right-click in the figure > Channels > Mark selected as bad Right-click on the "Link to raw file" > EEG > Display time series. For continuous MEG: ?Review continuous recordings and edit markers For continuous EEG: EEG and epilepsy For imported files: ?Exploring the recordings
In this experiment, the electric stimulation is sent with a frequency of 2.8Hz, meaning that the inter-stimulus interval is 357ms. We are going to import epochs of 300ms around the stimulation events. Right-click on the Link to raw file > Import in database: Check "Use events" and select "Trigger01" Epoch time: [-50, 250] ms Check "Remove DC offset" > Time range > [-50, -10] ms At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 250ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-50,250]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment, we can just ignore it.
License
Description of the experiment
Export recordings from Meg160
http://www.yokogawa.com/me/index.htm
(.txt file generally available under the corresponding "Scan" folder)
(DigitizeLabel.txt generally located in the "C:\Meg160\AppInfo" folder) Download and installation
Import the anatomy
Without the individual MRI
For more information, read the following tutorial: Warping default anatomy Access the recordings
Link the recordings
Prepare the channel file
Refine the MRI registration
Read the stimulation information
Artifacts: Evaluate the power spectrum
Evaluation
Electric contamination due to the power lines (60Hz+harmonics)
Electric noise coming from an unknown source Correction
Artifacts: Heartbeats and eye blinks [TO DO]
Review the recordings
Epoching and averaging
Import recordings
SCREEN CAPTURE
Answer Yes to this question to discard the last epoch. Average epochs
Source analysis
Head model
Noise covariance matrix
Inverse model
Z-score normalization
Regions of interest
Scripting
Graphic edition
Generate Matlab script
Feedback
