|
Size: 9768
Comment:
|
Size: 12943
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 11: Artifact detection = ''Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet'' |
= Tutorial 12: Artifact detection = ''Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet'' |
| Line 4: | Line 4: |
| The previous tutorial illustrated how to remove noise patterns occurring continuously and at specific frequencies. However, most of the events that contaminate the MEG/EEG recordings are not permanent, span over a large frequency range or overlap with the frequencies of the brain signals of interest. Frequency filters are not appropriate to correct for eye movements, breathing movements, heartbeats or other muscle activity. | The previous tutorials illustrated how to remove noise patterns occurring continuously and at specific frequencies. However, most of the events that contaminate the MEG/EEG recordings are not persistant, span over a large frequency range or overlap with the frequencies of the brain signals of interest. Frequency filters are not appropriate to correct for eye movements, breathing movements, heartbeats or other muscle activity. |
| Line 6: | Line 6: |
| Other approaches exist to correct for those artifacts, based on the spatial signature of the artifacts. If an event is very reproducible and occurs always at the same position (eg. eye blinks and heartbeats), the sensors will always record the same values when it occurs. We can identify the topographies corresponding to this artifact (ie spatial distributions of values at one time point) and remove them from the recordings. This spatial decomposition is the basic idea behind two widely used approaches: the '''SSP '''(Signal-Space Projection) and '''ICA '''(Independent Component Analysis) methods. We will describe those approaches in the next tutorial. The SSP method is based on the spatial decomposition of the MEG/EEG recordings for a selection of time samples during which the artifact is present. Therefore we need to identify when each type of artifact is occurring in the recordings. This tutorial shows how to detect automatically some well defined artifacts: the blinks and the heartbeats. |
For getting rid of reproducible artifacts, one popular approach is the Signal-Space Projection (SSP). This method is based on the spatial decomposition of the MEG/EEG recordings for a selection of time samples during which the artifact is present. Therefore we need to identify when each type of artifacts is occurring in the recordings. This tutorial shows how to automatically detect some well defined artifacts: the blinks and the heartbeats. |
| Line 20: | Line 18: |
| * On those traces, there is not much happening for most of the recordings except for a few bumps. This subject is sitting very still and not blinking much. We can expect MEG recordings of a very good quality. * '''ECG''': Right-click on the link > ECG > Display time series<<BR>>The electrocardiogram was recorded with a bipolar montage of electrodes across the chest. On the green trace, you can recognize the typical shape of the electric activity of the heart (P, QRS and T waves). |
* On these traces, there is not much happening for most of the recordings except for a few bumps. This subject is sitting very still and not blinking much. We can expect MEG recordings of a very good quality. * '''ECG''': Right-click on the link > ECG > Display time series.<<BR>>The electrocardiogram was recorded with a bipolar montage of electrodes across the chest. You can recognize the typical shape of the electric activity of the heart (P, QRS and T waves). |
| Line 24: | Line 22: |
| * To keep the scale fixed between to pages: Uncheck the button [AS] in the figure (auto-scale) | * To keep the scale fixed between two pages: Uncheck the button '''[AS]''' (auto-scale). |
| Line 27: | Line 25: |
| * The additional data channels (ECG and EOG) contain precious information that we will use for the automatic detection of the blinks and heartbeats. We strongly recommend that you always record these signals in your own experiments, it helps a lot with the data analysis. <<BR>><<BR>> {{attachment:observe.gif||height="271",width="662"}} | * The additional data channels (ECG and EOG) contain precious information that we can use for the automatic detection of the blinks and heartbeats. We strongly recommend that you always record these signals during your own experiments, it helps a lot with the data pre-processing. <<BR>><<BR>> {{attachment:observe.gif||height="271",width="662"}} |
| Line 30: | Line 28: |
| In the Record tab, select the menu: '''Artifacts > Detect heartbeats'''. | In the Record tab, select the menu: '''"Artifacts > Detect heartbeats"'''. |
| Line 32: | Line 30: |
| * It opens automatically the pipeline editor, with the process "Detect heartbeats" selected. * '''Channel name''': Name of the channel that is used to perform the detection. Select "'''ECG'''". * '''Time window''': Time range that the algorithm should scan for amplitude peaks. Leave the default values to process the entire file, or select the option '''[All file]'''. * '''Event name''': Name of the event group created for saving the detected events. Keep "'''cardiac'''".<<BR>><<BR>> {{attachment:detect_ecg.gif}} * Click on Run. After the process stops, you can see a new event category "'''cardiac'''". The 464 occurrences indicate a heart rate around 77bpm. * You can check a few of them, to make sure the "cardiac" markers really indicate the ECG peaks, and that there are not too many peaks that are skipped.<<BR>><<BR>> {{attachment:detect_ecg.gif}} |
* It automatically opens the pipeline editor, with the process "Detect heartbeats" selected. * '''Channel name''': Name of the channel that is used to perform the detection. Select or type "'''ECG'''". * '''Time window''': Time range that the algorithm should scan for amplitude peaks. Leave the default values to process the entire file, or check the option '''[All file]'''. * '''Event name''': Name of the event group created for saving the detected events. Enter "'''cardiac'''".<<BR>><<BR>> {{attachment:detect_ecg.gif||height="254",width="549"}} * Click on Run. After the process stops, you can see a new event category "'''cardiac'''". The 464 heartbeats for 360s of recordings indicate an average heart rate of 77bpm, everything looks normal. * You can check a few of them, to make sure the "cardiac" markers really indicate the ECG peaks. Not all peaks need to be detected, but you should have a minimum of 10-20 events marked for removing the artifacts using SSP, described in the following tutorials.<<BR>><<BR>> {{attachment:detect_ecg_done.gif||height="127",width="517"}} |
| Line 42: | Line 38: |
| Now do the same thing for the blinks. | Now do the same thing for the blinks: Menu "'''Artifacts > Detect eye blinks'''". |
| Line 44: | Line 40: |
| In the Record tab, select the menu: '''SSP > Detect eye blinks'''. It opens automatically the pipeline editor, with the process "Detect eye blinks" selected: | * Channel name: '''VEOG''' * Time window: '''All file''' * Event name: '''Blink''' <<BR>><<BR>> {{attachment:detect_eog.gif||height="257",width="550"}} * Run, then look quickly at the 15 detected blinks (shortcut: Shift+Right arrow). <<BR>><<BR>> {{attachment:detect_eog_done.gif||height="130",width="535"}} |
| Line 46: | Line 45: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspMenu.gif|sspMenu.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEog.gif|detectEog.gif|height="232",width="334",class="attachment"}} | == Remove simultaneous blinks/heartbeats == We will use these event markers as the input to our SSP cleaning method. This technique works well if each artifact is defined precisely and as independently as possible from the other artifacts. This means that we should try to avoid having two different artifacts marked at the same time. |
| Line 48: | Line 48: |
| Click on Run. After the process stops, you can see two new event categories "'''blink'''" and "'''blink2'''" in the Record tab. You can review a few of them, to make sure that they really indicate the EOG events. In the Record tab, click on the "blink" event category, then on a time occurrence to jump to it in the MEG and Misc time series figures. | Because the heart beats every second or so, there is a high chance that when the subject blinks there is a heartbeat not too far away in the recordings. We cannot remove all the blinks that are contaminated with a heartbeat because we would have no data left. But we have a lot of heartbeats, so we can do the contrary: remove the markers "cardiac" that are occurring during a blink. |
| Line 50: | Line 50: |
| Two types of events are created because this algorithm not only detects specific events in a signal, it also classifies them by shape. If you go through all the events that were detected in the two categories, you would see that the "blink" are all round bumps, typical of the '''eye blinks'''. In the category "blink2", the morphologies don't look as uniform; it mixes small blinks, and ramps or step functions followed by sharp drops that could indicate '''eye saccades'''. The saccades can be observed on the vertical EOG, but if you want a better characterization of them you should also record the horizontal EOG. The detection of the saccades should be performed with a different set of parameters, using the process "Detect custom events", introduced later in this chapter. | In the Record tab, select the menu "'''Artifacts > Remove ''''''simultaneous'''". Set the options: |
| Line 52: | Line 52: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEogDone.gif|detectEogDone.gif|class="attachment"}} | * Remove events named: "'''cardiac'''" * When too close to events: "'''blink'''" * Minimum delay between events: '''250ms''' <<BR>><<BR>> {{attachment:detect_simult.gif||height="257",width="550"}} |
| Line 54: | Line 56: |
| == Remove simultaneous blinks/hearbeats == SSP > Remove simultaneous > cardiac / blink / 250ms |
After executing this process, the number of "cardiac" events goes from '''464''' to '''455'''. The deleted heartbeats were all less than 250ms away from a blink. |
| Line 57: | Line 58: |
| == Running from a script == | == Run #02: Running from a script == |
| Line 60: | Line 61: |
| * Select the two AEF runs in the Process1 box. * Select successively the following processes, then click on [Run]: * '''Events > Detect heartbeats:''' Select channel '''ECG''', check "All file", event name "cardiac". * '''Events > Detect eye blinks:''' Select channel '''VEOG''', check "All file", event name "blink". * '''Events > Remove simultaneous''': Remove "'''cardiac'''", too close to "'''blink'''", delay '''250ms'''. |
* Close everything with the '''[X]''' button at the top-right corner of the Brainstorm window. * Select the run '''AEF #02''' in the Process1 box, then select the following processes: * '''Events > Detect heartbeats:''' Select channel '''ECG''', check "All file", event name "cardiac". * '''Events > Detect eye blinks:''' Select channel '''VEOG''', check "All file", event name "blink". * '''Events > Remove simultaneous''': Remove "'''cardiac'''", too close to "'''blink'''", delay '''250ms'''.<<BR>><<BR>> {{attachment:detect_script.gif||height="286",width="506"}} * Open the Run#02 recordings (MEG+EOG+ECG) and verify that the detection worked as expected. You should get '''472 cardiac''' events and '''19 blink''' events. |
| Line 68: | Line 70: |
| == Custom detection == Those two previous processes are shortcuts for a generic process "'''Detect custom events'''". We are not going to use it here, but it is interesting to introduce it to understand how the blinks and heartbeats detection work. The logic is the following: |
== Artifacts classification == If the EOG signals are not as clean as here, the detection processes may create more than one category, for instance: '''blink''', '''blink2''', '''blink3'''. The algorithm not only detects specific events in a signal, it also classifies them by shape. For two detected events, the signals around the event marker have to be sufficiently correlated (> 0.8) to be classified in the same category. At the end of the process, all the categories that contain less than 5 events are deleted. |
| Line 71: | Line 73: |
| * The channel to analyze is read from the continuous file, for a given time window. | In the good cases, this can provide an automatic classification of different types of artifacts, for instance: blinks, saccades and other eye movements. The tutorial [[Tutorials/MedianNerveCtf|MEG median nerve (CTF)]] is a good illustration of appropriate classification: '''blink''' groups the real blinks, and '''blink2''' contains mostly saccades. . {{attachment:detect_classification.gif||height="148",width="653"}} In the bad cases, the signal is too noisy and the classfication fails. It leads to either many different categories, or none if all the categories have less than 5 events. If you don't get good results with the process "Detect eye blinks", you can try to run a '''custom detection''' with the classification disabled. At the contrary, if you obtain one category that mixes multiple types of artifacts and would like to automatically separate them in different sub-groups, you can try the process "'''Events > Classify by shape'''". It is more powerful than the automatic classification from the event detection process because it can run on multiple signals at the same type: first it reduces the number of dimensions with a PCA decomposition, then runs a similar classification procedure. <<TAG(Advanced)>> == Detection: Custom events == These two processes "Detect heartbeats" and "Detect eye blinks" are in reality shortcuts for a generic process "'''Detect custom events'''". This process can be used for detecting any kind of event based on the signal power in a specific frequency band. We are not going to use it here, but you may have to use it if the standard parameters do not work well, or for detecting other types of events. * The signal to analyze is read from the continuous file (options "Channel name" and "Time window"). |
| Line 74: | Line 89: |
| * '''Minimum duration between two events''': If the filtered signal crosses the threshold several times in relation with the same artifact (like it would be the case for muscular activity recordings on an EMG channel), we don't want to trigger several events but just one at the beginning of the activity. This parameter would indicate the algorithm to take only the maximum value over the given time window; it also prevents from detecting other events immediately after a successful detection. For the ECG, this value is set to 500ms, because it is very unlikely that the heart rate of the subject goes over 120 beats per minute. | * '''Minimum duration between two events''': If the filtered signal crosses the threshold several times in relation with the same artifact (eg. muscle activity in an EMG channel), we don't want to trigger several events but just one at the beginning of the activity. This parameter would indicate the algorithm to take only the maximum value over the given time window; it also prevents from detecting other events immediately after a successful detection. For the ECG, this value is set to 500ms, because it is very unlikely that the heart rate of the subject goes over 120 beats per minute. |
| Line 76: | Line 91: |
| * '''Enable classification''': If this option is selected, the events are classified by shape, based on correlation measure. In the end, only the categories that have more than 5 occurrences are kept, all the other successful detections are ignored. | * '''Enable classification''': If this option is selected, the events are classified by shape in different categories, based on correlation measure. In the end, only the categories that have more than 5 occurrences are kept, all the other successful detections are ignored. <<BR>><<BR>> {{attachment:detect_custom.gif||height="435",width="313"}} |
| Line 78: | Line 93: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectCustom.gif|detectCustom.gif|height="459",width="329",class="attachment"}} | <<TAG(Advanced)>> |
| Line 80: | Line 95: |
| === In case of failure === If you cannot get your artifacts to be detected automatically, you can browse through the recordings and mark all the artifacts manually, as explained in the previous tutorial [[Tutorials/EventMarkers|Event markers]]. |
== In case of failure == If the signals are not as clean as in this sample dataset, the automatic detection of the heartbeats and blinks may fail with the standard parameters. You may have to use the process "Detect custom events" and adjust some parameters. For instance: |
| Line 83: | Line 98: |
| <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials/ArtifactsFilter&next=Tutorials/ArtifactsSsp")>> | * If nothing is detected: decrease the amplitude threshold, or try to adjust the frequency band. * If too many events are detected: increase the amplitude threshold or the minimum duration between two events. * If too many categories of events are generated, and you have a very little number of events in the end: disable the classification. * To find the optimal frequency band for an artifact, you can open the recordings and play with the online band-pass filters in the Filter tab. Keep the band that shows the highest amplitude peaks. If you cannot get your artifacts to be detected automatically, you can browse through the recordings and mark all the artifacts manually, as explained in the tutorial [[Tutorials/EventMarkers|Event markers]]. == Other detection processes == * Events >''' Detect analog trigger''': See tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/StimDelays#Detection_of_the_analog_triggers|Stimulation delays]] * Events > '''Detect events above threshold''': Trigger events based on amplitude levels. * Events > '''Detect other artifacts''': See tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/BadSegments|Additional bad segments]] * Events > '''Detect movement''': See tutorial [[Tutorials/MovementDetect|Detect subject movements]] * Synchronize > '''Transfer events''': See tutorial [[Tutorials/EyetrackSynchro|Synchronization with eye tracker]] * Artifacts > '''Detect bad channels: Peak-to-peak''': Reject channels and trials from imported data. == Additional documentation == * Tutorial: [[Tutorials/MovementDetect|Detect subject movements]] * Tutorial: [[Tutorials/EyetrackSynchro|Synchronization with eye tracker]] <<HTML(<!-- END-PAGE -->)>> <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials/BadChannels&next=Tutorials/ArtifactsSsp")>> |
Tutorial 12: Artifact detection
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
The previous tutorials illustrated how to remove noise patterns occurring continuously and at specific frequencies. However, most of the events that contaminate the MEG/EEG recordings are not persistant, span over a large frequency range or overlap with the frequencies of the brain signals of interest. Frequency filters are not appropriate to correct for eye movements, breathing movements, heartbeats or other muscle activity.
For getting rid of reproducible artifacts, one popular approach is the Signal-Space Projection (SSP). This method is based on the spatial decomposition of the MEG/EEG recordings for a selection of time samples during which the artifact is present. Therefore we need to identify when each type of artifacts is occurring in the recordings. This tutorial shows how to automatically detect some well defined artifacts: the blinks and the heartbeats.
Contents
Observation
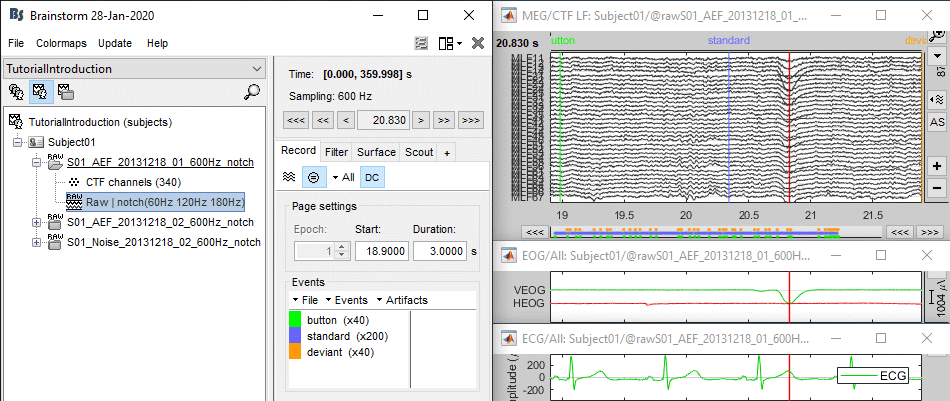
Let's start by observing the type of contamination the blinks and heartbeats cause to the MEG recordings.
Run #01: Double-click on the link to show the MEG sensors.
Configuration: Page of 3 seconds, view in columns, selection of the "CTF LT" sensors (the left-temporal sensors will be a good example to show at the same time the two types of artifacts).
EOG: Right-click on the link > EOG > Display time series. Two channels are classified as EOG:
VEOG: Vertical electrooculogram (two electrodes placed below and above one eye)
HEOG: Horizontal electrooculogram (two electrodes placed on the temples of the subject)
- On these traces, there is not much happening for most of the recordings except for a few bumps. This subject is sitting very still and not blinking much. We can expect MEG recordings of a very good quality.
ECG: Right-click on the link > ECG > Display time series.
The electrocardiogram was recorded with a bipolar montage of electrodes across the chest. You can recognize the typical shape of the electric activity of the heart (P, QRS and T waves).Find a blink: Scroll through the recordings using the F3 shortcut until you see a large blink.
- Remember you can change the amplitude scale with many shortcuts (eg. right-click + move).
To keep the scale fixed between two pages: Uncheck the button [AS] (auto-scale).
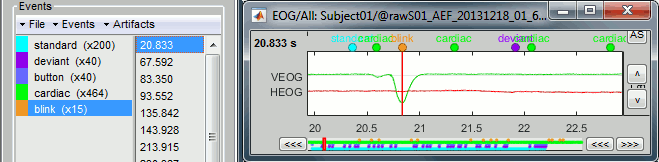
- For instance, you can observe a nice blink at 20.8s (red cursor in the screen capture below).
- On the same page, you should be able to observe the contamination due to a few heartbeats, corresponding to the peaks of the ECG signal (eg. 19.8s, shown as a blue selection below).
The additional data channels (ECG and EOG) contain precious information that we can use for the automatic detection of the blinks and heartbeats. We strongly recommend that you always record these signals during your own experiments, it helps a lot with the data pre-processing.
Detection: Heartbeats
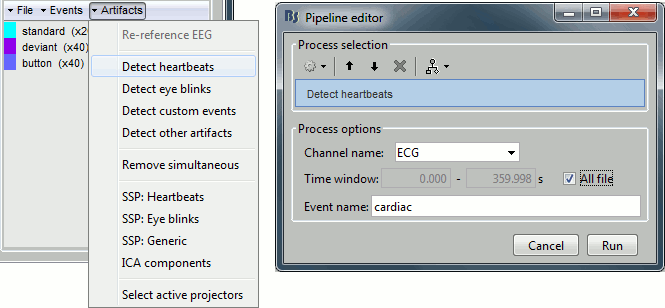
In the Record tab, select the menu: "Artifacts > Detect heartbeats".
- It automatically opens the pipeline editor, with the process "Detect heartbeats" selected.
Channel name: Name of the channel that is used to perform the detection. Select or type "ECG".
Time window: Time range that the algorithm should scan for amplitude peaks. Leave the default values to process the entire file, or check the option [All file].
Event name: Name of the event group created for saving the detected events. Enter "cardiac".
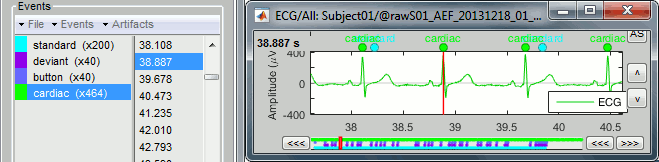
Click on Run. After the process stops, you can see a new event category "cardiac". The 464 heartbeats for 360s of recordings indicate an average heart rate of 77bpm, everything looks normal.
You can check a few of them, to make sure the "cardiac" markers really indicate the ECG peaks. Not all peaks need to be detected, but you should have a minimum of 10-20 events marked for removing the artifacts using SSP, described in the following tutorials.
Detection: Blinks
Now do the same thing for the blinks: Menu "Artifacts > Detect eye blinks".
Channel name: VEOG
Time window: All file
Event name: Blink
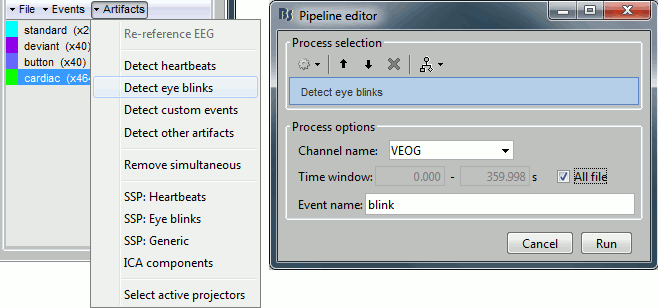
Run, then look quickly at the 15 detected blinks (shortcut: Shift+Right arrow).
Remove simultaneous blinks/heartbeats
We will use these event markers as the input to our SSP cleaning method. This technique works well if each artifact is defined precisely and as independently as possible from the other artifacts. This means that we should try to avoid having two different artifacts marked at the same time.
Because the heart beats every second or so, there is a high chance that when the subject blinks there is a heartbeat not too far away in the recordings. We cannot remove all the blinks that are contaminated with a heartbeat because we would have no data left. But we have a lot of heartbeats, so we can do the contrary: remove the markers "cardiac" that are occurring during a blink.
In the Record tab, select the menu "Artifacts > Remove simultaneous". Set the options:
Remove events named: "cardiac"
When too close to events: "blink"
Minimum delay between events: 250ms
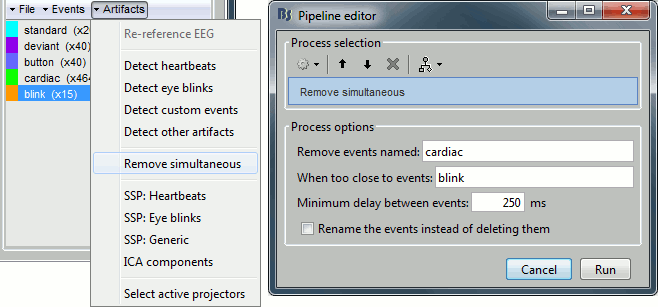
After executing this process, the number of "cardiac" events goes from 464 to 455. The deleted heartbeats were all less than 250ms away from a blink.
Run #02: Running from a script
Let's perform the same detection operations on Run #02, using this time the Process1 box.
Close everything with the [X] button at the top-right corner of the Brainstorm window.
Select the run AEF #02 in the Process1 box, then select the following processes:
Events > Detect heartbeats: Select channel ECG, check "All file", event name "cardiac".
Events > Detect eye blinks: Select channel VEOG, check "All file", event name "blink".
Events > Remove simultaneous: Remove "cardiac", too close to "blink", delay 250ms.
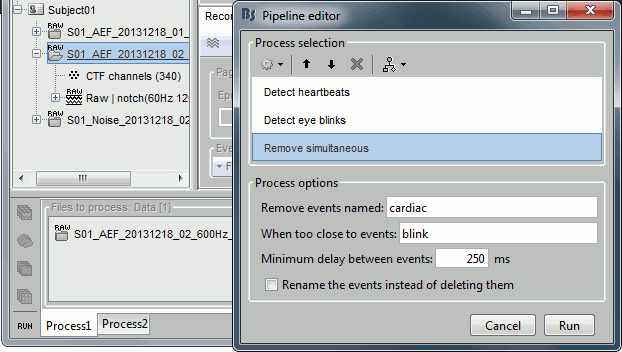
Open the Run#02 recordings (MEG+EOG+ECG) and verify that the detection worked as expected. You should get 472 cardiac events and 19 blink events.
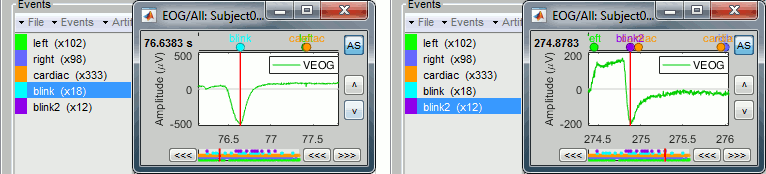
Artifacts classification
If the EOG signals are not as clean as here, the detection processes may create more than one category, for instance: blink, blink2, blink3. The algorithm not only detects specific events in a signal, it also classifies them by shape. For two detected events, the signals around the event marker have to be sufficiently correlated (> 0.8) to be classified in the same category. At the end of the process, all the categories that contain less than 5 events are deleted.
In the good cases, this can provide an automatic classification of different types of artifacts, for instance: blinks, saccades and other eye movements. The tutorial MEG median nerve (CTF) is a good illustration of appropriate classification: blink groups the real blinks, and blink2 contains mostly saccades.
In the bad cases, the signal is too noisy and the classfication fails. It leads to either many different categories, or none if all the categories have less than 5 events. If you don't get good results with the process "Detect eye blinks", you can try to run a custom detection with the classification disabled.
At the contrary, if you obtain one category that mixes multiple types of artifacts and would like to automatically separate them in different sub-groups, you can try the process "Events > Classify by shape". It is more powerful than the automatic classification from the event detection process because it can run on multiple signals at the same type: first it reduces the number of dimensions with a PCA decomposition, then runs a similar classification procedure.
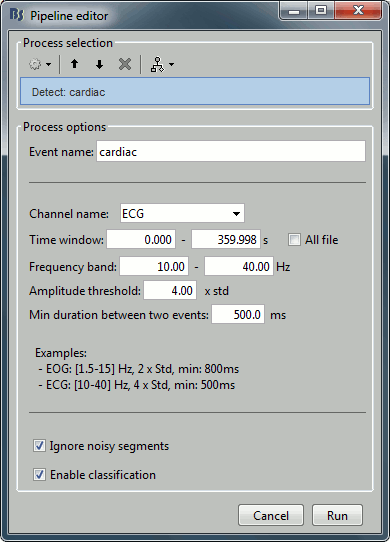
Detection: Custom events
These two processes "Detect heartbeats" and "Detect eye blinks" are in reality shortcuts for a generic process "Detect custom events". This process can be used for detecting any kind of event based on the signal power in a specific frequency band. We are not going to use it here, but you may have to use it if the standard parameters do not work well, or for detecting other types of events.
- The signal to analyze is read from the continuous file (options "Channel name" and "Time window").
Frequency band: The signal is filtered in a frequency band where the artifact is easy to detect. For EOG: 1.5-15Hz ; for ECG: 10-40Hz.
Threshold: An event of interest is detected if the absolute value of the filtered signal value goes over a given number of times the standard deviation. For EOG: 2xStd, for ECG: 4xStd
Minimum duration between two events: If the filtered signal crosses the threshold several times in relation with the same artifact (eg. muscle activity in an EMG channel), we don't want to trigger several events but just one at the beginning of the activity. This parameter would indicate the algorithm to take only the maximum value over the given time window; it also prevents from detecting other events immediately after a successful detection. For the ECG, this value is set to 500ms, because it is very unlikely that the heart rate of the subject goes over 120 beats per minute.
Ignore the noisy segments: If this option is selected, the detection is not performed on the segments that are much noisier than the rest of the recordings.
Enable classification: If this option is selected, the events are classified by shape in different categories, based on correlation measure. In the end, only the categories that have more than 5 occurrences are kept, all the other successful detections are ignored.
In case of failure
If the signals are not as clean as in this sample dataset, the automatic detection of the heartbeats and blinks may fail with the standard parameters. You may have to use the process "Detect custom events" and adjust some parameters. For instance:
- If nothing is detected: decrease the amplitude threshold, or try to adjust the frequency band.
- If too many events are detected: increase the amplitude threshold or the minimum duration between two events.
- If too many categories of events are generated, and you have a very little number of events in the end: disable the classification.
- To find the optimal frequency band for an artifact, you can open the recordings and play with the online band-pass filters in the Filter tab. Keep the band that shows the highest amplitude peaks.
If you cannot get your artifacts to be detected automatically, you can browse through the recordings and mark all the artifacts manually, as explained in the tutorial Event markers.
Other detection processes
Events > Detect analog trigger: See tutorial Stimulation delays
Events > Detect events above threshold: Trigger events based on amplitude levels.
Events > Detect other artifacts: See tutorial Additional bad segments
Events > Detect movement: See tutorial Detect subject movements
Synchronize > Transfer events: See tutorial Synchronization with eye tracker
Artifacts > Detect bad channels: Peak-to-peak: Reject channels and trials from imported data.
Additional documentation
Tutorial: Detect subject movements
Tutorial: Synchronization with eye tracker