Brainstorm-FieldTrip auditory tutorial
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Elizabeth Bock, Robert Oostenveld.
The aim of this tutorial is to provide high-quality recordings of a simple auditory stimulation and illustrate the best analysis paths possible with Brainstorm and FieldTrip. This page presents the workflow in the Brainstorm environment, the equivalent documentation for the FieldTrip environment will be available on the FieldTrip website.
Note that the operations used here are not detailed, the goal of this tutorial is not to teach Brainstorm to a new inexperienced user. For in depth explanations of the interface and the theory, please refer to the 12+3 introduction tutorials.
Contents
License
This tutorial dataset (EEG and MRI data) remains a property of the MEG Lab, McConnell Brain Imaging Center, Montreal Neurological Institute, McGill University, Canada. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the MEG Lab.
If you reference this dataset in your publications, please aknowledge its authors (Elizabeth Bock, Peter Donhauser, Francois Tadel and Sylvain Baillet) and cite Brainstorm as indicated on the website. For questions, please contact us through the forum.
Presentation of the experiment
Experiment
- One subject
- One acquisition run = 200 regular beeps + 40 easy deviant beeps
- Two runs of 6 minutes each
- Random inter-stimulus interval: between 0.7s and 1.7s seconds, uniformly distributed
- The subject presses a button when detecting a deviant
- Auditory stim generated with the Matlab Psychophysics Toolbox
MEG acquisition
- Acquisition at 2400Hz, with a CTF 275 system at the MNI, subject in seating position
- Online 600Hz low-pass filter, files saved with the 3rd order gradient
- Recorded Channels: ...
- 3 datasets:
S01_AEF_20131218_01: run1 - marked a few trials bad => Use for the main example
S01_AEF_20131218_02: run2 - saccades, see component screenshot, they are not really detected well with the auto detection => To illustrate how to remove saccades. External file of marked saccades
- Noise recordings: 30s
- File name: S01 = Subject01, AEF = Auditory evoked field, 20131218 = date Dec 18 2013, 01=run
Use of the .ds, not the AUX (standard at the MNI) because they are easier to manipulate in FieldTrip
Head shape and fiducial points
3D digitization using a Polhemus Fastrak device driven by Brainstorm (http://neuroimage.usc.edu/brainstorm/Tutorials/TutDigitize)
- The output file is copied to each .ds folder and contains the following entries:
- the position of the center of CTF coils
- the position of the anatomical references we use in Brainstorm (nasion and connections tragus/helix - the red point I placed on that ear image we have on both websites)
- Around 150 head points distributed on the hard parts of the head (no soft tissues)
Subject anatomy
- Subject with 1.5T MRI
- Marker on the left cheek
Processed with FreeSurfer 5.3
Download and installation
- Requirements: You have already followed all the basic tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: sample_auditory.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself.
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialAuditory" and select the options:
"No, use individual anatomy",
"No, use one channel file per condition".
Import the anatomy
- Switch to the "anatomy" view.
Right-click on the TutorialAuditory folder > New subject > Subject01
- Leave the default options you set for the protocol
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: sample_auditory/anatomy
- Number of vertices of the cortex surface: 15000 (default value)
- Set the 6 required fiducial points (indicated in MRI coordinates):
- NAS: x=127, y=213, z=139
- LPA: x=52, y=113, z=96
- RPA: x=202, y=113, z=91
- AC: x=127, y=119, z=149
- PC: x=128, y=93, z=141
- IH: x=131, y=114, z=206 (anywhere on the midsagittal plane)
- At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.
Access the recordings
Link the recordings
- Switch to the "functional data" view.
Right-click on the subject folder > Review raw file
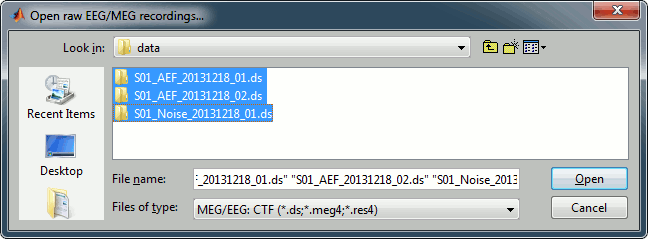
Select the file format: "MEG/EEG: CTF (*.ds...)"
Select all the .ds folders in: sample_auditory/data
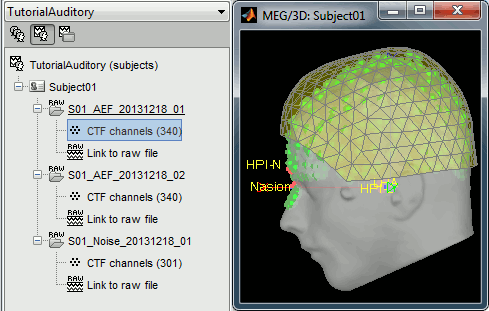
Refine registration now? YES
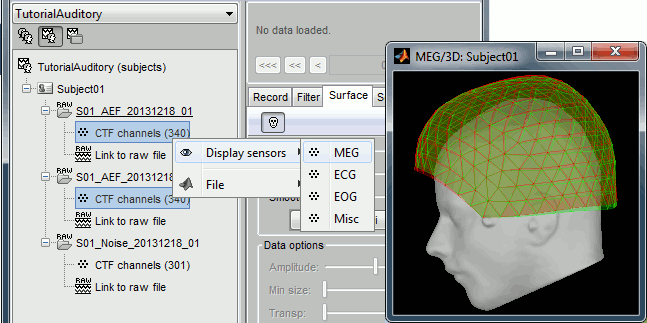
Multiple runs and head position
The two AEF runs 01 and 02 were acquired successively, the position of the subject's head in the MEG helmet was estimated twice, once at the beginning of each run. The subject might have moved between the two runs. To evaluate visually the displacement between the two runs, select at the same time all the channel files you want to compare (the ones for run 01 and 02), right-click > Display sensors > MEG.
- Typically, we would like to group the trials coming from multiple runs by experimental conditions. However, because of the subject's movements between runs, it's not possible to directly compare the sensor values between runs because they probably do not capture the brain activity coming from the same regions of the brain.
- You have three options if you consider grouping information from multiple runs:
- Method 1: Process all the runs separately and average between runs at the source level: The more accurate option, but requires a lot more work, computation time and storage.
- Method 2: Ignore movements between runs: This can be acceptable for commodity if the displacements are really minimal, less accurate but much faster to process and easier to manipulate.
Method 3: Co-register properly the runs using the process Standardize > Co-register MEG runs: Can be a good option for displacements under 2cm. Warning: This method has not be been fully evaluated on our side, to use at your own risk.
- In this tutorial, we will illustrate only method 1: runs are not co-registered.
Epoched vs. continuous
- The CTF MEG system can save two types of files: epoched (.ds) or continuous (_AUX.ds).
- Here we have an intermediate storage type: continuous recordings saved in an "epoched" file. The file is saved as small blocks of recordings of a constant time length (1 second in this case). All those time blocks are contiguous, there is no gap between them.
- Brainstorm can consider this file either as a continuous or an epoched file. By default it imports the regular .ds folders as epoched, but we can change this manually, to process it as a continuous file.
Double-click on the "Link to raw file" for run 01 to view the MEG recordings. You can navigate in the file by blocks of 1s, and switch between blocks using the "Epoch" box in the Record tab. The events listed are relative to the current epoch.
Right-click on the "Link to raw file" for run 01 > Switch epoched/continuous
Double-click on the "Link to raw file" again. Now you can navigate in the file without interruptions. The box "Epoch" is disabled and all the events in the file are displayed at once.
- Repeat this operation twice to convert all the files to a continuous mode.
Run 02 > Switch epoched/continuous
Noise > Switch epoched/continuous
Detect and remove artifacts
Spectral evaluation
- One of the typical pre-processing steps consist in getting rid of the contamination due to the power lines (50 Hz or 60Hz). Let's start with the spectral evaluation of this file.
- Drag all the "Link to raw file" to the Process1 box, or easier, just drag the node "Subject01", it will select recursively all the files in it.
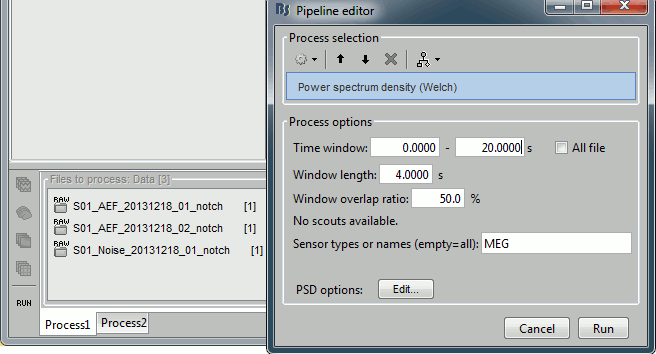
Run the process "Frequency > Power spectrum density (Welch)". Configure it as illustrated in the following figure (window length=10s, overlap=50%).
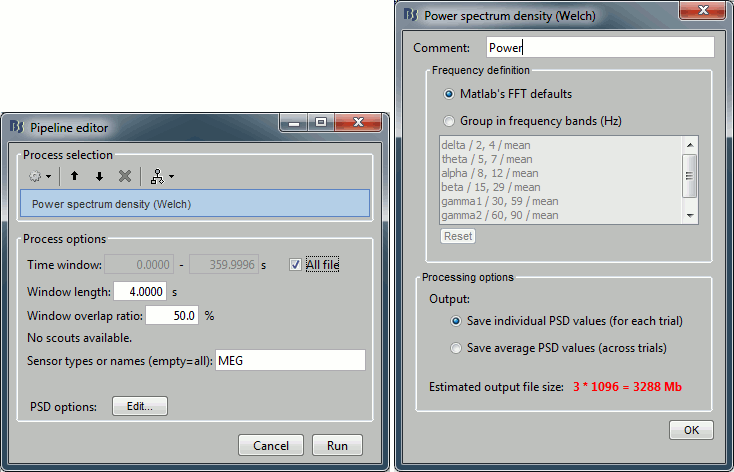
Double-click on the new PSD files to display them.
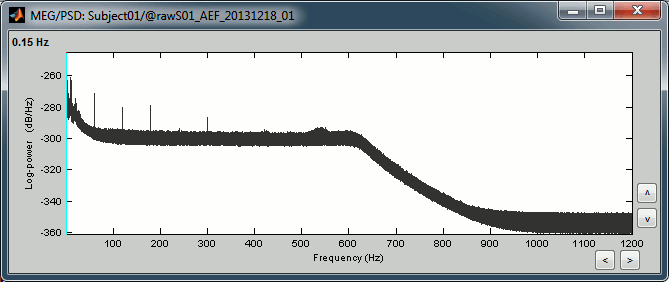
- We can observe 3 clear peaks related with the power lines: 60Hz, 120Hz, 180Hz
- The decrease of power after 600Hz corresponds to the low-pass filter applied at the acquisition time.
Power line contamination
- Put all the "Link to raw file" into the Process1 box (or the Subject01 node)
Run the process: Pre-process > Sinusoid removal (notch)
- Select the frequencies: 60, 120, 180 Hz
- This will take a while...
In output, this process creates new .ds folders in the same folder as the original files, and links the new files to the database.
{{attachment:psd3.gif}
Run again the PSD on those new files to evaluate the quality of the correction
=> SCREEN CAPTURE: psd4.gifTo avoid the confusion, delete the links to original files: select the folders containing the original files (not the new ones) and press the Delete key.
Cardiac artifact
cardiac SSP only 11% but I would use it, see MLT sensors, see component screenshot
run1 - marked a few trials bad
run2 - saccades, see component screenshot, they are not really detected well with the auto detection
Epoching and averaging
import standard and deviant: [-100,500]ms, remove DC with [-100,0]ms baseline
- do not make new condition - keep trials separate according to run average trial types see screen shot of MEG and EEG - P300 can be seen here even with only 40 epochs in the average
Source estimation
Head model
headmodel overlapping spheres, used cortex 15000 vertices
Noise covariance matrix
Noise recordings
Inverse model
sources min norm
z-score with baseline [-100,0] sec, abs of sources
Regions of interest
scouts over primary auditory cortex (A1)
see screen shots
Time-frequency
TF over scout 1, z-scored [-100,0] sec
see screen shots interesting gamma in the deviant
Discussion
Discussion about the choice of the dataset on the FieldTrip bug tracker:
http://bugzilla.fcdonders.nl/show_bug.cgi?id=2300
Scripting
Process selection
Graphic edition
Generate Matlab script
This list of processes can be converted automatically to a Matlab script.
- See the results on the page:
Feedback
