|
Size: 4247
Comment:
|
Size: 6830
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| ''Authors: ***'' | ''Authors: Chinmay Chinara, Samuel Medina Villalon, Takfarinas Medani, Raymundo Cassani, Anand Joshi, Christian Bénar, Richard Leahy'' |
| Line 13: | Line 13: |
| <<TableOfContents(3,2)>> | <<TableOfContents(2,2)>> |
| Line 23: | Line 23: |
| * GARDEL [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|brainstorm plugin]] needs to be installed for doing the automatic detection of the contacts. | * GARDEL [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|brainstorm plugin]] needs to be installed for doing the automatic detection of the contacts. If you are using Brainstorm from MATLAB, GARDEL requires the [[https://www.mathworks.com/products/image-processing.html|Image Processing Toolbox]]. The [[https://www.mathworks.com/products/parallel-computing.html|Parallel Computing Toolbox]] is recommended but not required. |
| Line 28: | Line 28: |
| * In the iEEG panel, click on '''Electrodes > Automatic localization > GARDEL'''. Click '''Yes''' on the disclaimer that pops up.<<BR>><<BR>> {{attachment:auto_gardel_menu.png||width="450"}} * In few minutes, you can see the electrodes being rendered live on the figures. While GARDEL is a great tool but there could be some bad detections due to factors mentioned in the disclaimer. It is always good to verify the results carefully. * If there are any bad detections, they can be post-processed using editing features available within Brainstorm. More details can be found [[https://neuroimage.usc.edu/brainstorm/seeg/SeegContactLocalization#Edit_the_contacts_positions|here]]. |
* In the iEEG panel, click on '''Electrodes > Automatic localization > GARDEL'''. Click '''Yes''' on the disclaimer that pops up.<<BR>><<BR>> {{attachment:auto_gardel_menu.png||width="450"}} <<BR>><<BR>> {{attachment:2_disclaimer.png||width="350"}} * In few minutes, you can see the electrodes being rendered live on the figures.<<BR>><<BR>> {{attachment:1_gardel_detect.png||width="650"}} <<BR>><<BR>> <<HTML(<center><iframe width="560" height="315" src="https://www.youtube.com/embed/rkixWwRkUS0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe></center>)>> |
| Line 32: | Line 31: |
| '''TODO: Show some bad detections with this tutorial data and steps how to correct them here''' '''TODO: Add relevant screenshots''' |
== Recommendations == * While GARDEL is a great tool but there could be some bad detections (that can be seen above) due to factors mentioned in the disclaimer. It is always good to verify the results carefully. * This is a particularly complex case and it can be seen that there are that there are a lot of overlapping and concentrated electrodes that are mostly touching and they are difficult to separate. GARDEL does fail to detect them and would require manual correction. It can very well be achieved using the editing features available within Brainstorm (check the [[https://neuroimage.usc.edu/brainstorm/seeg/SeegContactLocalization#Edit_the_contacts_positions|Edit the contact positions]] section). * Another way to achieve better detection is to work in the CT space because the resolution of CT is much higher than that of MRI. To do that, import the CT first, set the fiducials in the CT, import MRI, coregister MRI to CT and then run SPM skull stripping and generating Isosurface from that. Running GARDEL on this would give a much better result. Natively, GARDEL actually recommends working in the CT space. Please refer to the paper below for more details. * A video showing the automatic detection of contacts in the CT space using the GARDEL's sample data as provide in their [[https://gitlab-dynamap.timone.univ-amu.fr/public_shared_tools/gardel/-/wikis/home|website]] can be seen as under.<<BR>><<BR>> <<HTML(<center><iframe width="560" height="315" src="https://www.youtube.com/embed/PHi4YgiOwDU" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe></center>)>> |
| Line 37: | Line 38: |
| === Forum discussions === | |
| Line 39: | Line 39: |
| * S. Medina Villalon, R. Paz, N. Toehri, S. Lagarde, F. Pizzo, B. Colombet, F. Bartolomei, R. Carron, C.-G. Bénar. [[https://doi.org/10.1016/j.jneumeth.2018.03.018|EpiTools, A software suite for presurgical brain mapping in epilepsy: Intracerebral EEG]]. <<BR>> Journal of Neuroscience Methods 2018. |
|
| Line 40: | Line 42: |
| * [[https://neuroimage.usc.edu/brainstorm/seeg/Introduction|CT to MRI co-registration]] * [[https://neuroimage.usc.edu/brainstorm/seeg/SeegContactLocalization|Contact localization and labeling]] * [[https://neuroimage.usc.edu/brainstorm/seeg/SeizureFingerprinting|Time-Frequency Fingerprint Analysis for Epileptogenic Zone Localization]] * [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity|Epileptogenicity]] |
Automatic SEEG Contact Localization using GARDEL
Authors: Chinmay Chinara, Samuel Medina Villalon, Takfarinas Medani, Raymundo Cassani, Anand Joshi, Christian Bénar, Richard Leahy
This tutorial describes how to perform automatic SEEG contact localization in Brainstorm using GARDEL.
Please note that this tutorial is intended for users already familiar with Brainstorm. It does not provide detailed explanations of the software's interface or theoretical foundations.
For comprehensive introductory material, refer to the Brainstorm introduction tutorials.
Make sure you complete the CT to MRI co-registration and Contact localization and labeling advanced tutorial before proceeding any further.
NOT FOR CLINICAL USE:
The methods and software implementations presented in this tutorial have not been certified as medical devices. They are intended for research purposes only and should not be used for clinical decision-making.
Contents
Introduction
GARDEL (GUI for Automatic Registration and Depth Eectrode Localization) was developed by a multidisciplinary team as part of the EpiTools suite, led by Samuel Medina Villalon (Aix‑Marseille University & AP‑HM, Marseille), Rodrigo Manuel Paz (University of Buenos Aires–CONICET), Nicolas Roehri, Stanislas Lagarde, Fabrice Pizzo, Bruno Colombet, Fabrice Bartolomei, Romain Carron, and Christian Bénar, all contributing through INSERM and partnering institutions across France, Argentina, and Switzerland. This international group combined clinical neurophysiology and advanced imaging expertise to create GARDEL, enabling fully automatic detection and anatomical labeling of SEEG contacts thereby laying a strong foundation for streamlining SEEG analysis in research workflows.
Requirements
Complete the CT to MRI co-registration to have the data and the required anatomy set up.
Follow the tutorial Contact localization and labeling to generate the isosurface from the CT with a threshold of 2400 Hounsefiled Unit. This threshold plays a key part in the automatic detection as the the better segmented the electrodes are the better is the detection. After much trial and error, 2400 threshold gave the best results.
GARDEL brainstorm plugin needs to be installed for doing the automatic detection of the contacts. If you are using Brainstorm from MATLAB, GARDEL requires the Image Processing Toolbox. The Parallel Computing Toolbox is recommended but not required.
Automatic contact localization using GARDEL
Right click on Subject01, and choose SEEG/ECOG implantation.
Choose MRI+CT+IsoSurf from the popup menu (more details about the menu can be found here). While any of the options can be chosen from the menu, this gives the option to visualize and interact both in 3D and 2D.
In the iEEG panel, click on Electrodes > Automatic localization > GARDEL. Click Yes on the disclaimer that pops up.
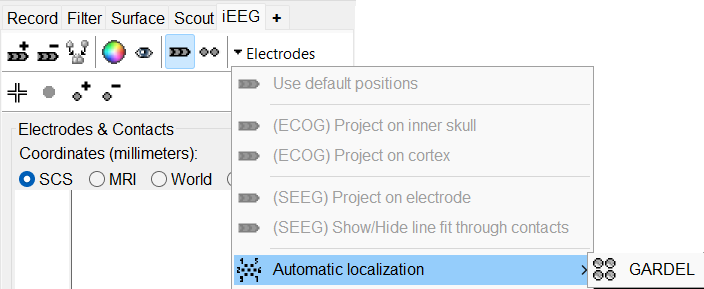
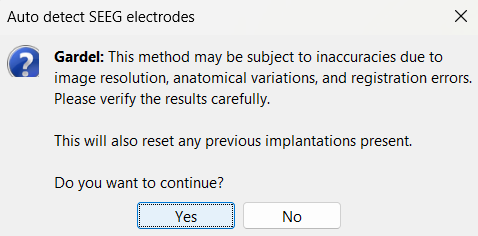
In few minutes, you can see the electrodes being rendered live on the figures.
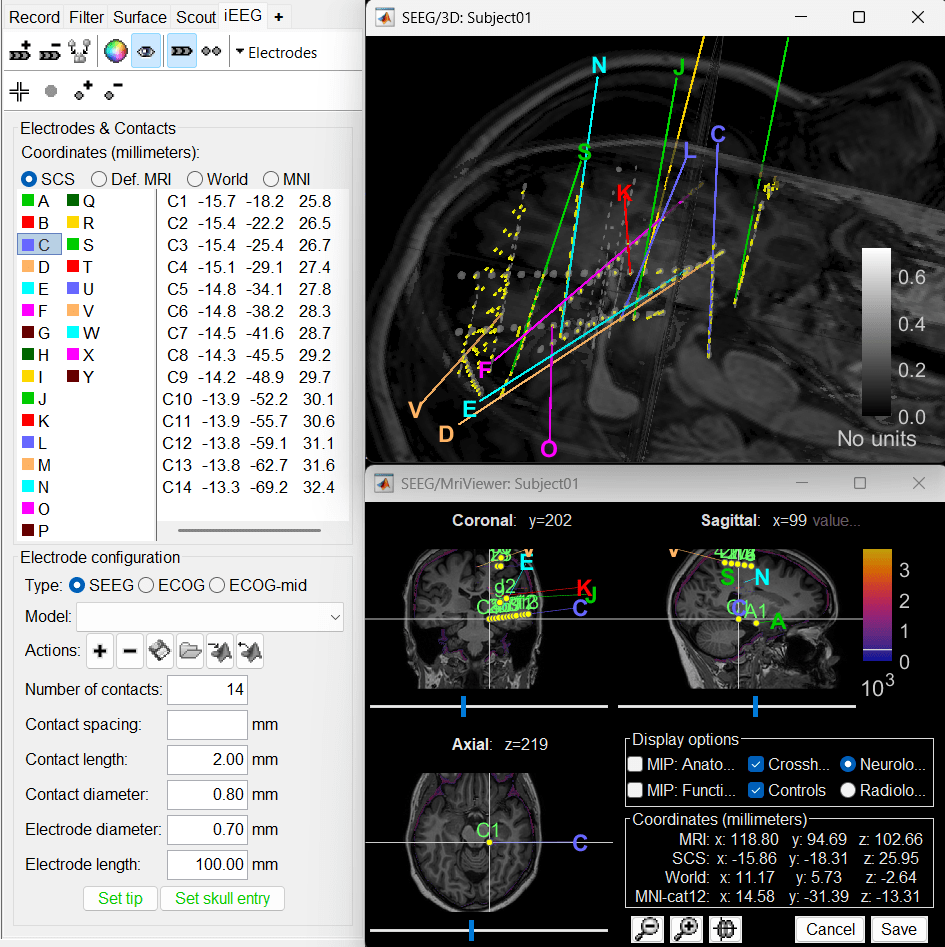
Recommendations
- While GARDEL is a great tool but there could be some bad detections (that can be seen above) due to factors mentioned in the disclaimer. It is always good to verify the results carefully.
This is a particularly complex case and it can be seen that there are that there are a lot of overlapping and concentrated electrodes that are mostly touching and they are difficult to separate. GARDEL does fail to detect them and would require manual correction. It can very well be achieved using the editing features available within Brainstorm (check the Edit the contact positions section).
- Another way to achieve better detection is to work in the CT space because the resolution of CT is much higher than that of MRI. To do that, import the CT first, set the fiducials in the CT, import MRI, coregister MRI to CT and then run SPM skull stripping and generating Isosurface from that. Running GARDEL on this would give a much better result. Natively, GARDEL actually recommends working in the CT space. Please refer to the paper below for more details.
A video showing the automatic detection of contacts in the CT space using the GARDEL's sample data as provide in their website can be seen as under.
Additional Documentation
Articles
S. Medina Villalon, R. Paz, N. Toehri, S. Lagarde, F. Pizzo, B. Colombet, F. Bartolomei, R. Carron, C.-G. Bénar. EpiTools, A software suite for presurgical brain mapping in epilepsy: Intracerebral EEG.
Journal of Neuroscience Methods 2018.
