Using the anatomy templates
Author: Francois Tadel
Contents
Changing the default anatomy
When you create a new protocol, the program makes a copy of the ICBM152 anatomy (previously Colin27) and sets it as the default for the protocol. It means that you will be able to use this template brain as a substitute for the subjects without an individual MRI, or as the common brain for group analysis.
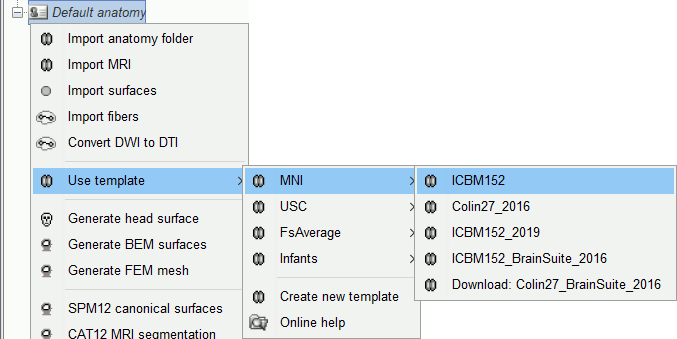
Other sets of MRI+surfaces are available to replace the ICBM152 anatomy. Right-click on (Default anatomy) > Use template. If a package is not currently available on your system, it will be downloaded from the Brainstorm website and saved in $HOME/.brainstorm/templates.
If you click on any of the download options, it downloads it into your templates folder, then the list of files in the (default anatomy) folder is replaced with the new template.
If the automatic download doesn't work, you can download the templates manually from the Download page and copy the .zip files directly in the folder $HOME/.brainstorm/templates.
Group analysis
When performing a group analysis with multiple subjects for which you have the individual MRI scans, you need to project the sources estimated on each subject on a common template, as explained in this tutorial: Group analysis.
For accurate registration between different brains (from a subject to a template or between subjects), you need to use a template that was generated using the same program as the one you used for running the segmentation of all the subjects of your studies.
You can use either BrainSuite or FreeSurfer for processing the MRIs or your subjects, but you need to use a template that matches this choice in order to use the accurate registration methods.
FreeSurfer templates
Available options:
Colin27_2016: Average of 27 scans of the same head, FreeSurfer 5.3: more information
ICBM152_2016: Non-linear average of 152 subjects, FreeSurfer 5.3: more information
ICBM152: Distributed directly with the Brainstorm package. Same as ICBM152_2016, but without the white matter envelopes, in order to minimize the size of the standard Brainstorm distribution.
FSAverage_2016: Average of 40 subjects using a spherical averaging (Fischl et al. 1999).
It is the default FreeSurfer brain: please register here if you are using it.
More images: http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#FSAverage_template
They all include the following information:
- T1 MRI volume
- Cortex/white surface: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices)
- Head layers: scalp, outer skull, inner skull
FreeSurfer spherical registration of each hemisphere, for subject co-registration.
FreeSurfer surface-based atlases: Desikan-Killiany, Destrieux, Brodmann, Mindboggle
(plus Yeo2011 and PALS for FSAverage only)- ASEG sub-cortical atlas.
For more information on the interactions between FreeSurfer and Brainstorm: read this tutorial.
BrainSuite templates
Available options:
Colin27_BrainSuite_2016: Average of 27 scans, processed with BrainSuite 15b: more information
ICBM152_BrainSuite_2016: Non-linear average of 152 subjects, BrainSuite 15b: more information
BCI-DNI_BrainSuite_2020: Single subject atlas from USC, BrainSuite 15c: more information
USCBrain_BrainSuite_2020: Anatomical and functional hybrid atlas from USC, BrainSuite 17a: more
Warning: If you are using BrainSuiteAtlas1 for BrainSuite processing, then you should use Colin27_BrainSuite_2016 or ICBM152_BrainSuite_2016 as the default anatomy. If you are using the BCI-DNI_brain_atlas, then you should use BCI-DNI_BrainSuite_2016 as the template in BrainStorm.
They all include the following information:
- T1 MRI volume
- Cortex/white surfaces: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices)
- Head layers: scalp, outer skull, inner skull
BrainSuite square registration of each hemisphere, for subject co-registration.
BrainSuite surface-based atlas: SVReg
For more information on the interactions between BrainSuite and Brainstorm: read this tutorial.
BrainVISA templates
Note that the BrainVISA-based templates do not allow any accurate registration procedure.
Colin27_2012: Previous version of the default anatomy distributed with Brainstorm.
Infant7w_2015b: 7-week infant brain with the anatomical atlas from (Kabdebon et al. 2014).
Oreilly_1y: 1 year old infant brain with Tzourio-Mazoyer surface atlas (Li et al. 2015, Shi et al. 2011): http://neuroimage.usc.edu/forums/showthread.php?2123-Atlas-for-1-year-old-babies
Modify the default MRI fiducials
The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm. By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot in the Coordinates systems page.
If you want to use an anatomy template but you are using a different convention when digitizing the position of these points, you have to modify the default positions of the template with the MRI Viewer.
- Go to the anatomy view
In (default anatomy), right-click on the MRI > Edit MRI
- Modify the position of the fiducial points to match your own convention
- Click on [Save], it will update the surfaces to match the new coordinate system