|
Size: 6860
Comment:
|
Size: 14109
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Realistic head model: FEM with Duneuro = '''[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]''' |
= Realistic head model: FEM with DUNEuro = '''[TUTORIAL UNDER REVISION]''' |
| Line 4: | Line 4: |
| ''Authors: Takfarinas, Juan, Francois, Sophie, Maria, Johannes, Christian, Carsten, John, Richard ? '' | ''Authors: [[https://neuroimage.usc.edu/brainstorm/AboutUs/tmedani|Takfarinas Medani]], Juan Garcia-Prieto, Francois Tadel, Sophie Schrader, Anand Joshi, Christian Engwer, Carsten Wolters, John Mosher and Richard Leahy '' |
| Line 6: | Line 6: |
| This tutorial explains how to use brainstorm-dueneuro to compute the head model using the finite element method, implemented in the Duneuro software. | {{attachment:logo_duneuro.png||align="right",height="82",width="187"}} |
| Line 8: | Line 8: |
| This forward model uses a finite element element method (FEM) and was developed by the '''...(l''''''ink)'''. It uses the volume mesh of the realistic head model. The goal of this forward solution is mostly for '''EEG users''', to provide more accurate results than the spherical models and more complex geometry than the BEM. It is not necessary for MEG users, as the "overlapping spheres" method gives similar results but much faster. This method is illustrated using the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG and epilepsy]] or similar data ('''todo'''). | This tutorial explains how to use [[http://duneuro.org/|DUNEuro]] to compute the forward model using the '''finite element method''' ('''FEM'''). The FEM methods use a realistic volume mesh of the head generated from the segmentation of the MRI. The FEM models provide more accurate results than the spherical forward models and more realistic geometry and tissue properties than the [[Tutorials/TutBem|BEM]] methods. |
| Line 10: | Line 10: |
| refe to these point : http://duneuro.org/ | The scope of this page is limited to a '''basic example''' (head model with 3 layers). More advanced options for mesh generation and forward model computation are discussed in other tutorials: [[Tutorials/FemMesh|FEM mesh generation]], [[Tutorials/FemTensors|FEM tensors estimation]], [[Tutorials/FemMedianNerve|FEM median nerve example]]. We assume that you have already followed the [[Tutorials|introduction tutorials]], we will not discuss the general principles of forward modeling here. |
| Line 12: | Line 12: |
| and to the documentation from here : | <<TableOfContents(2,2)>> |
| Line 14: | Line 14: |
| https://gitlab.dune-project.org/duneuro/duneuro/wikis/home | == DUNEuro == '''[[http://duneuro.org/|DUNEuro]] '''is an open-source C++ software library for solving partial differential equations (PDE) in neurosciences using mesh-based methods. It is based on the''' [[https://www.dune-project.org/|DUNE library]] '''and its main features include solving the EEG and MEG forward problem and providing simulations for brain stimulation. |
| Line 16: | Line 17: |
| Here we will describe the FEM and run some examples | As distributed on the [[http://gitlab.dune-project.org/duneuro/duneuro|DUNEuro GitLab]], the source code works only on Linux operating systems. Interfaces to Matlab and Python are possible, but you need to install and compile DUNEuro by yourself. For Brainstorm, we adapted this code and were able to generate '''binaries''' for the main operating systems''' '''(Windows, '''Linux '''and MacOS), which are '''downloaded automatically''' when needed as a [[Tutorials/Plugins|Brainstorm plugin]]. This project is available on our [[https://github.com/brainstorm-tools/bst-duneuro|GitHub repository bst-duneuro]]. |
| Line 18: | Line 19: |
| == Duneuro == DUNEuro is an open-source C++ software library for solving partial differential equations in neurosciences using mesh bases methods. It is based on the DUNE library and its main features include solving the electroencephalography (EEG) and magnetoencephalography (MEG) forward problem and providing simulations for brain stimulation. |
We would like to thank the''' DUNEuro team '''for their help with this integration work: Carsten Wolters, Christian Engwer, Sophie Schrader, Andreas Nuessing, Tim Erdbruegger, Marios Antonakakis, Johannes Vorwerk & Maria Carla Piastra. |
| Line 21: | Line 21: |
| http://duneuro.org/ | ''' {{attachment:duneuroFromDune.JPG||width="546",height="187"}} ''' Please '''cite the following papers''' if you use this software in your work: * Medani T, Garcia-Prieto J, Tadel F, Schrader S, Antonakakis M, Joshi A, Engwer C, Wolters CH, Mosher JC, Leahy RM, [[https://doi.org/10.1117/12.2580935|Realistic head modeling of electromagnetic brain activity: an integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions]] ([[http://www.sci.utah.edu/~wolters/PaperWolters/2021/MedaniEtAl_SPIE_2021.pdf|preprint]]), SPIE Medical Imaging (2021) Publications that make use of, or extend the DUNEuro library (listed on the [[http://duneuro.org/|duneuro website]]): * Piastra MC, Nüßing A, Vorwerk J, Bornfleth H, Oostenveld R, Engwer C, Wolters CH, [[https://www.frontiersin.org/articles/10.3389/fnins.2018.00030/full|The Discontinuous Galerkin Finite Element Method for Solving the MEG and the combined MEG/EEG Forward Problem]], Frontiers in Neuroscience (2018) * Engwer C, Vorwerk J, Ludewig J, Wolters CH, [[https://epubs.siam.org/doi/abs/10.1137/15M1048392|A discontinuous Galerkin method to solve the EEG forward problem using the subtraction approach]], SIAM Journal on Scientific Computing (2017) * Nüßing A, Wolters CH, Brinck H, Engwer C, [[https://ieeexplore.ieee.org/document/7511781|The unfitted discontinuous Galerkin method for solving the EEG forward problem]], IEEE Transactions on Biomedical Engineering (2016)''' ''' |
| Line 24: | Line 34: |
| Duneuro is build on top of DUNE Library, at this time (September 2019) the source code works only on Linux operating systems. However we are able to generate the binaries for the main platforms (windows 64, Linux and Mac), therefor it can be easily used from Matlab for all platforms without the need to install and compile the tidiuos CPP library of DUNE and DUNEURO. | In order to reproduce the computation present below on your computer, you need to fulfill all the conditions listed below. Alternatively, you can read this page as a reference documentation about DUNEuro and not try to reproduce the results. |
| Line 26: | Line 36: |
| However if you want to use the source code of Duneuro you can visite: http://duneuro.org/ | * You have a working copy of Brainstorm installed on your computer. * You have already followed the [[https://neuroimage.usc.edu/brainstorm/Introduction|introduction tutorials]], at least until #23. * You have followed the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG and Epilepsy]], as it is used for illustrating the computation. * '''DUNEuro''': Software installed automatically as a [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|Brainstorm plugin]]. * '''Iso2mesh''': Software installed automatically as a [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|Brainstorm plugin]]. |
| Line 28: | Line 42: |
| For the advanced user users, he main steps you need in order to compile for windows are listed here : https://github.com/svdecomposer/brainstorm-duneuro | == FEM mesh == In order to use the FEM computations of the electromagnetic field (EEG/MEG), the volume mesh of the head is required. Brainstorm integrates most of the modern open-source tools to generate realistic head mesh, either from nested surface mesh or from individual MR images (T1 or T1 and T2). This tutorial describes only a simple approach based on three nested surfaces meshed in 3D with Iso2mesh. For the full list of available methods and description of options, please refer to the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh|FEM mesh generation]].''' ''' |
| Line 30: | Line 45: |
| We distribute the binary files for Windows/Ubunto 64bit systems, you don't need to installan extra Microsoft software package to run '''Duneuro.''' | * Select the protocol TutorialEpilepsy, created while following the tutorial [[http://Tutorials/Epilepsy|EEG and epilepsy]]. * Go to the anatomy view. * Select the three [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#BEM_surfaces|BEM surfaces]]: scalp, outer skull, inner skull. * Right-click > '''Generate FEM mesh''' > Iso2mesh-2021 > MergeMesh > Default options. <<BR>><<BR>> {{attachment:femMesh1.gif}} <<BR>> {{attachment:iso2mesh.gif}} ''' ''' * The FEM mesh appears in the database explorer after a short while. The first number indicates the number of vertices (i.e. nodes) of the tetrahedral mesh. To get the number of 3D elements (i.e. tetrahedrons) in this geometric model of the head: right-click on the file > File > View file contents. The structure of the file is describe in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#On_the_hard_drive|FEM mesh generation]].<<BR>><<BR>> {{attachment:femMesh2.gif}} * Double-click on the FEM file to display it. From the Surface tab, you can change the resection locations by moving the bottom sliders, and you can control the display of each of the three layers individually. Click on the layer button in the toolbar, then adjust the color and transparency of the corresponding mesh. The figure below represents the FEM mesh in the center and the initial BEM layers on the right. <<BR>><<BR>> {{attachment:femMesh3.gif}} |
| Line 32: | Line 52: |
| '''<<TAG(Advanced)>>''' | == FEM forward model == The '''forward model''' (or '''head model''' in the Brainstorm documentation and interface) describes how the electric activity in the '''source space''' (the cortex surface or a regular grid of volume points) influences the electric potential (EEG) or magnetic fields (MEG) at the level of the '''sensors'''. The FEM method uses the '''tetrahedral mesh''' computed above to establish this relationship. ''' ''' |
| Line 34: | Line 55: |
| However, to use this pipline from Brainstorm you need to download the bst-duneuro toolbox (ref) that interface brainstorm to duneuro. | * Go the Anatomy view. Select the default FEM mesh and cortex surface you'd like to use for the computation (in case there is more than one in each category). The selected elements appear in green, double-click or right-click > Set as default to change the selection. * Go to the Functional view. Right-click on the channel file > '''Compute head model'''. <<BR>><<BR>> {{attachment:femCompute1.gif}} * Select '''MRI volume''', EEG: '''DUNEuro FEM''', select all the layers and the default conductivity values (scalp = inside the head surface, skull = inside the outer skull surface, brain = inside the inner skull surface).<<BR>><<BR>> {{attachment:femCompute2.gif}} * Volume source grid: '''Regular grid''', brain, 5mm. {{attachment:femCompute3.gif}} * Start the computation. Depending on the complexity of the problem (number of FEM elements, number of layers, number of source points, number of sensors and computation options), this can take anywhere between a few minutes to a few days. In this example, if you selected the correct files and options, it should only take a few minutes. You may be prompted to download or update the bst-duneuro [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|plugin]]. <<BR>><<BR>> {{attachment:femCompute4.gif}} |
| Line 36: | Line 61: |
| The FEM mesh visualisation and some mesh processing requires the installation of the iso2mesh toolbox (link). If you want to work on offline, you can download it and add it to your matlab path. If it's not installed, Brainstrom will download and istall this toolbox when it needed. | == Source analysis == When it's finished, you have now a new head model for this subject in your database and can be used for the next steps/[[https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel|(back to tutorial 20 : Head modeling)]] |
| Line 38: | Line 64: |
| == FEM surfaces / Volume generation == === Volume generation from surface files === In this part you can generate your FEM mesh from surfaces that you can get fron the segmentation software (brainSuite, FreeSurfer ....). |
<<TAG(Advanced)>> |
| Line 42: | Line 66: |
| This process will | == DUNEuro options: Basic == When assuming '''isotropic conductivities''' for all the tissues, the DUNEuro basic options options are limited to the selection of the tissues and their conductivities. |
| Line 44: | Line 69: |
| - merge the surfaces, | . {{attachment:duneuroBasic.gif}} {{attachment:duneuroTensors.gif}} |
| Line 46: | Line 71: |
| - check the self intersecting | * '''FEM layers: '''Brainstorm reads the lists of tissues from the selected FEM mesh. Users can select the desired layers to include in the FEM computation. According to the modalities, the recommended selections are: * '''EEG''': Select all the layers * '''MEG''': Select only the inner layers (inside the inner skull = brain = white+gray+csf) * '''SEEG''': Select only the inner layers * '''ECOG''': Select only the inner layers * Any combination of modalities that includes MEG: select all the layers |
| Line 48: | Line 78: |
| - fixe the size of the mesh | * '''FEM conductivities (isotropic):''' Left figure - Brainstorm proposes default conductivity values for each layer (derived from the SimBio software, see [[https://www.mrt.uni-jena.de/simbio/images/VorwerkChoRamppHamerKnoescheWolters_NeuroImage_2014_Webversion.pdf|this article]]). However, you have the possibility to change these values according to your model. |
| Line 50: | Line 80: |
| - generate the volum mesh | * '''FEM conductivities (anisotropic)''': Right figure - Advanced users can use information from '''diffusion imaging''' exams to estimate anisotropic '''conductivity tensors''' for each element of the FEM mesh. For more information, refer to the tutorial: [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemTensors|FEM tensors estimation]]. To remove previously computed tensors and restore the default isotropic approach, right-click on the FEM mesh > Clear FEM tensors. <<BR>><<BR>> {{attachment:clearFemTensors.JPG||width="220",height="300"}} |
| Line 52: | Line 82: |
| - visual checking ... | == DUNEuro options: Advanced [TODO] == In the DUNEuro options panel, click on the button '''Show details''' to access the advanced options. |
| Line 54: | Line 85: |
| - TODO : may be we can add some mesh auqlity measures ?? | . {{attachment:duneuroAdvanced.gif}} |
| Line 56: | Line 87: |
| '''<<TAG(Advanced)>>''' | * '''FEM layers & conductivities''': Discussed in the previous section. |
| Line 58: | Line 89: |
| === Volume generation from T1/T2 MRI data === You can also generate your own FEM head model and then load it to brainstorm. However the automatic head model generation from from imaging techniques are not accurate and most of the time visual checking are needed and manual correction are required. |
* '''FEM solver type''': (only the fitted FEM are integrated in Brainstorm at the moment) * '''CG''': Continuous Galerkin: This is the standard Lagrangian method. |
| Line 61: | Line 92: |
| ==> this depends lagely on the quality of the T1/T2 MRI image(https://simnibs.github.io/simnibs/build/html/tutorial/head_meshing.html). | * '''DC''': Discontinuous Galerkin: |
| Line 63: | Line 94: |
| This step is based on the "roast" toolbox (link to roast : https://github.com/andypotatohy/roast | * '''FEM source model''': * '''Venant''': * Number of moments: * Reference length: * Weighting exponent: * Relaxation factor exponent: * Mixed moment: * Restrict: * '''Subtraction''': * intorderadd: * intorderadd_lb: * '''Partial integration''': * '''Source space''': * '''Shrink source space''': The dipoles are moved inward by the specified distance (in mm). * '''Force source space''': This is required when the dipoles are not within the gray matter. * '''Output options''': * '''Save transfer matrix''': |
| Line 65: | Line 112: |
| ) that we adapted for the MEEG forward computation. If you want to generate your own FEM head model from an MRI, you will need to download these file (link), then run the bst process as explained here. | For more information about these methods, users can check [[https://www.sci.utah.edu/~wolters/PaperWolters/2016/Vorwerk_Dissertation_2016.pdf|Johannes Vorwerk's PhD thesis]]. |
| Line 67: | Line 114: |
| === FEM Head model generation with SimNibs === This method used the SimNibs software. So to call this process, you need to download and install the SimNibs software, the process of the installation is explained in the SimNibs webpage : https://simnibs.github.io/simnibs/build/html/installation/simnibs_installer.html. |
== Troubleshooting [TODO] == DUNEuro binaries may crash for various reasons: we tried to list the possible causes here. Many FEM forward modeling issues are related with memory overload or extremely long computation times. Reducing the size of the problem may help in many cases. If you cannot find a solution, please post the full error message on the Brainstorm user forum (you can copy-paste the error message from the Matlab command window after closing the error message box). |
| Line 70: | Line 117: |
| When you have installed SimNibs, Brainstorm can call the main function used for the mesh generation frm the main graphical interface. Depemding on your computer performances, this process will take between 2 to 5 hours. We highly recommend to close all other running process and application on our computer in order to speed this process. | === Grid error messages === * Error message: Coordinate is outside of the grid, or grid is not convex * Explanation: Some dipoles are probably outside of the cortex, users need to correct the source space. * Solution: ? |
| Line 72: | Line 122: |
| - Explain here the main steps with screenshots : | === Remove the neck === One way to reduce the size of the forward problem is to decrease the number of FEM elements in the head model. When the field of the MRI is large, you may have the mesh of the neck and even the shoulders. In most cases, it is safe to remove the lower part of the FEM mesh, below the nose and the brainstem. Right-click on the FEM mesh > '''Resect Neck'''. |
| Line 74: | Line 125: |
| 1- Create new subject within the current protocole | {{attachment:resectNeck.JPG||width="600",height="150"}} |
| Line 76: | Line 127: |
| 2- Load the T1 of the subject to the brainstorm database. | Once the process is finished, a new FEM mesh appears in the database, with a tag "resect". The following figure shows the model before (743828 vertices / 4079587 elements) and after resection (613955 vertices / 3400957 elements). It will reduce the size of the problem by 20%. |
| Line 78: | Line 129: |
| 3- Associate a T2 mri to the subject if it's available (this is better for csf/skull/scalp segmentation) 4- Right click on the subject, select the "Generate FEM mesh" . Select "SIMNIBS", and choose "Tetrahedral element" and keep the other options to the default value. 5- When this process is finished, a new node will appear in the data base, which hase he name "FEM xxxV, (simNibs, yLayers)". This is the FEM mesh model generated from the T1. === FEM Head model template === - Load the FEM volumic mesh (template created from ICBM T1 MRI using SimNibs) - Load the surface mesh (template created also from ICBM using ICBM ) and then generates the volume mesh (either tetra or hexa) by calling the tetgen process cia iso2mesh toolbox (if hexa are desired, the tetra mesh will be converted to hexa ... ) === Head model based on the level set approach === TODO and Validate == FEM computation and unterface to DUNEuro == === Head model === Number of layers, conductivity value, isotropy/anisotropy/ mesh resolution/ === Electrode model === Check the position of the electrodes and align to the head model (projection if needed) === Source model === Similarly to the spherical nad BEM head model, the source position are defined on the cortex surface vertices. We can either define a contraned or not constrained orientation. However, for the FEM model, more paramters could be tuned for the source model. Choice of the source model : PI, Venant, Subtraction, Whitney Panel of the options choice that the user can select. (other wise we will set to default ) '''<<TAG(Advanced)>>''' === Advanced paramaters === - Solver parameters - Electrodes projection - maybe explain here the relevant option of the mini file ?? |
{{attachment:FemMeshAllandResect.JPG||width="600",height="150"}} |
| Line 120: | Line 132: |
| refer to : | ==== Related tutorials ==== * [[Tutorials/FemMesh|FEM mesh generation]] * [[Tutorials/FemTensors|FEM tensors estimation]] * [[Tutorials/FemMedianNerve|FEM median nerve example]] |
| Line 122: | Line 137: |
| http://duneuro.org/ | ==== DUNEuro references ==== * DUNEuro wiki: https://gitlab.dune-project.org/duneuro/duneuro/wikis/home * DUNEuro website: http://duneuro.org/ * List of the parameters: https://docs.google.com/spreadsheets/d/1MqURQsszn8Qj3-XRX_Z8qFFnz6Yl2-uYALkV-8pJVaM/edit#gid=0 |
| Line 124: | Line 142: |
| https://www.dune-project.org/ == Reported Errors & alternative solution == '''<<TAG(Advanced)>>''' == The MEEG forward problem with the FEM == == License == == Reference == |
==== Brainstorm-DUNEuro integration ==== * https://github.com/brainstorm-tools/bst-duneuro/issues/1 * https://github.com/brainstorm-tools/brainstorm3/issues/242 * https://github.com/brainstorm-tools/brainstorm3/issues/185 |
Realistic head model: FEM with DUNEuro
[TUTORIAL UNDER REVISION]
Authors: Takfarinas Medani, Juan Garcia-Prieto, Francois Tadel, Sophie Schrader, Anand Joshi, Christian Engwer, Carsten Wolters, John Mosher and Richard Leahy

This tutorial explains how to use DUNEuro to compute the forward model using the finite element method (FEM). The FEM methods use a realistic volume mesh of the head generated from the segmentation of the MRI. The FEM models provide more accurate results than the spherical forward models and more realistic geometry and tissue properties than the BEM methods.
The scope of this page is limited to a basic example (head model with 3 layers). More advanced options for mesh generation and forward model computation are discussed in other tutorials: FEM mesh generation, FEM tensors estimation, FEM median nerve example. We assume that you have already followed the introduction tutorials, we will not discuss the general principles of forward modeling here.
Contents
DUNEuro
DUNEuro is an open-source C++ software library for solving partial differential equations (PDE) in neurosciences using mesh-based methods. It is based on the DUNE library and its main features include solving the EEG and MEG forward problem and providing simulations for brain stimulation.
As distributed on the DUNEuro GitLab, the source code works only on Linux operating systems. Interfaces to Matlab and Python are possible, but you need to install and compile DUNEuro by yourself. For Brainstorm, we adapted this code and were able to generate binaries for the main operating systems (Windows, Linux and MacOS), which are downloaded automatically when needed as a Brainstorm plugin. This project is available on our GitHub repository bst-duneuro.
We would like to thank the DUNEuro team for their help with this integration work: Carsten Wolters, Christian Engwer, Sophie Schrader, Andreas Nuessing, Tim Erdbruegger, Marios Antonakakis, Johannes Vorwerk & Maria Carla Piastra.
Please cite the following papers if you use this software in your work:
Medani T, Garcia-Prieto J, Tadel F, Schrader S, Antonakakis M, Joshi A, Engwer C, Wolters CH, Mosher JC, Leahy RM, Realistic head modeling of electromagnetic brain activity: an integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions (preprint), SPIE Medical Imaging (2021)
Publications that make use of, or extend the DUNEuro library (listed on the duneuro website):
Piastra MC, Nüßing A, Vorwerk J, Bornfleth H, Oostenveld R, Engwer C, Wolters CH, The Discontinuous Galerkin Finite Element Method for Solving the MEG and the combined MEG/EEG Forward Problem, Frontiers in Neuroscience (2018)
Engwer C, Vorwerk J, Ludewig J, Wolters CH, A discontinuous Galerkin method to solve the EEG forward problem using the subtraction approach, SIAM Journal on Scientific Computing (2017)
Nüßing A, Wolters CH, Brinck H, Engwer C, The unfitted discontinuous Galerkin method for solving the EEG forward problem, IEEE Transactions on Biomedical Engineering (2016)
Requirements
In order to reproduce the computation present below on your computer, you need to fulfill all the conditions listed below. Alternatively, you can read this page as a reference documentation about DUNEuro and not try to reproduce the results.
- You have a working copy of Brainstorm installed on your computer.
You have already followed the introduction tutorials, at least until #23.
You have followed the tutorial EEG and Epilepsy, as it is used for illustrating the computation.
DUNEuro: Software installed automatically as a Brainstorm plugin.
Iso2mesh: Software installed automatically as a Brainstorm plugin.
FEM mesh
In order to use the FEM computations of the electromagnetic field (EEG/MEG), the volume mesh of the head is required. Brainstorm integrates most of the modern open-source tools to generate realistic head mesh, either from nested surface mesh or from individual MR images (T1 or T1 and T2). This tutorial describes only a simple approach based on three nested surfaces meshed in 3D with Iso2mesh. For the full list of available methods and description of options, please refer to the tutorial FEM mesh generation.
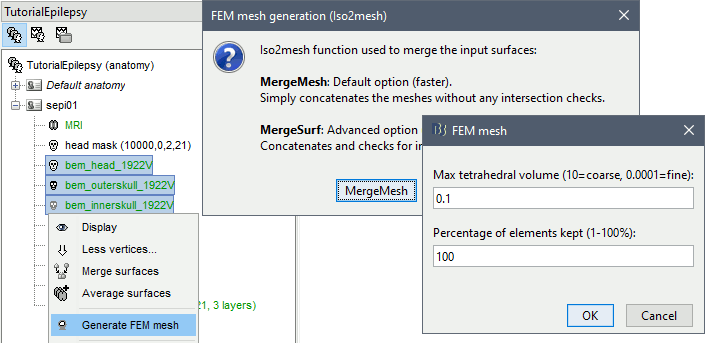
Select the protocol TutorialEpilepsy, created while following the tutorial EEG and epilepsy.
- Go to the anatomy view.
Select the three BEM surfaces: scalp, outer skull, inner skull.
Right-click > Generate FEM mesh > Iso2mesh-2021 > MergeMesh > Default options.
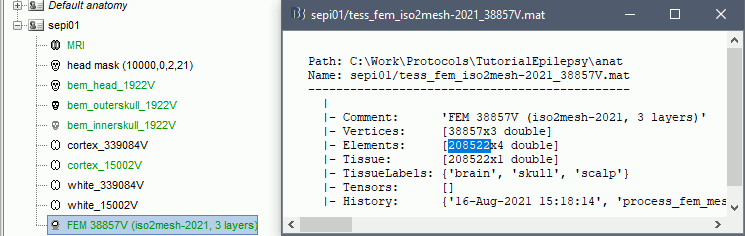
The FEM mesh appears in the database explorer after a short while. The first number indicates the number of vertices (i.e. nodes) of the tetrahedral mesh. To get the number of 3D elements (i.e. tetrahedrons) in this geometric model of the head: right-click on the file > File > View file contents. The structure of the file is describe in the tutorial FEM mesh generation.
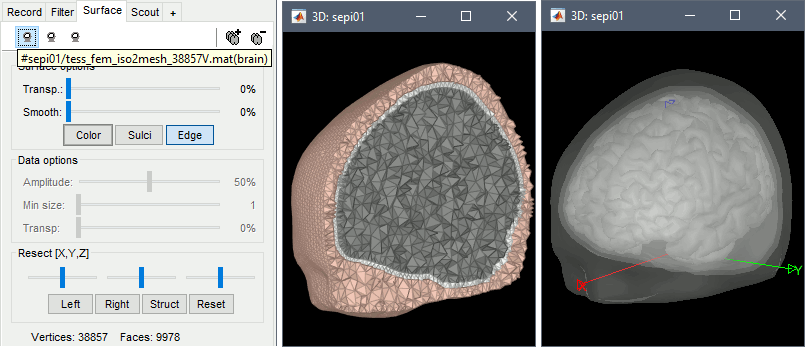
Double-click on the FEM file to display it. From the Surface tab, you can change the resection locations by moving the bottom sliders, and you can control the display of each of the three layers individually. Click on the layer button in the toolbar, then adjust the color and transparency of the corresponding mesh. The figure below represents the FEM mesh in the center and the initial BEM layers on the right.
FEM forward model
The forward model (or head model in the Brainstorm documentation and interface) describes how the electric activity in the source space (the cortex surface or a regular grid of volume points) influences the electric potential (EEG) or magnetic fields (MEG) at the level of the sensors. The FEM method uses the tetrahedral mesh computed above to establish this relationship.
Go the Anatomy view. Select the default FEM mesh and cortex surface you'd like to use for the computation (in case there is more than one in each category). The selected elements appear in green, double-click or right-click > Set as default to change the selection.
Go to the Functional view. Right-click on the channel file > Compute head model.
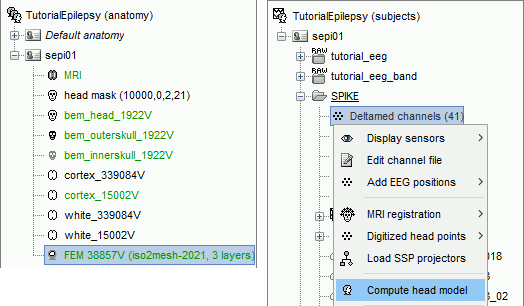
Select MRI volume, EEG: DUNEuro FEM, select all the layers and the default conductivity values (scalp = inside the head surface, skull = inside the outer skull surface, brain = inside the inner skull surface).
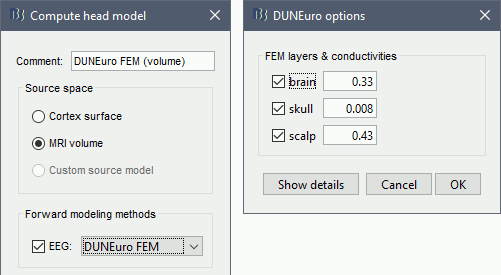
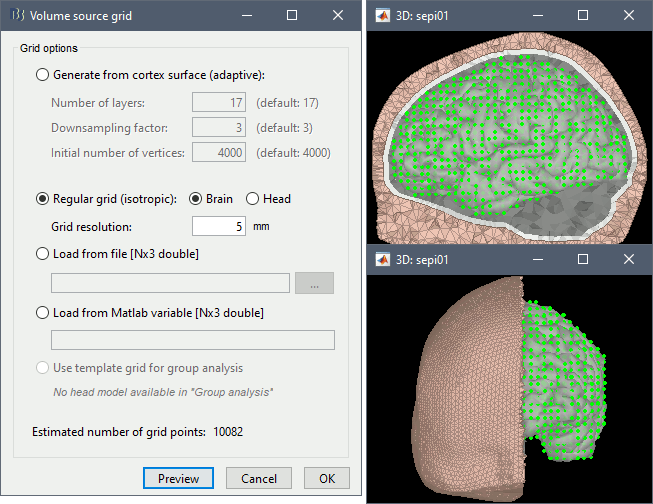
Volume source grid: Regular grid, brain, 5mm.
Start the computation. Depending on the complexity of the problem (number of FEM elements, number of layers, number of source points, number of sensors and computation options), this can take anywhere between a few minutes to a few days. In this example, if you selected the correct files and options, it should only take a few minutes. You may be prompted to download or update the bst-duneuro plugin.
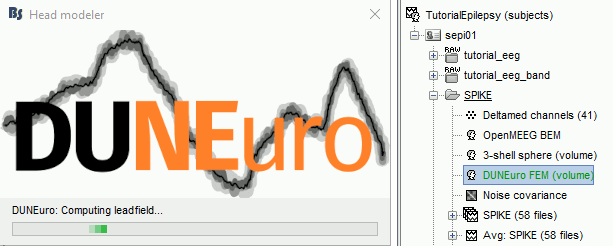
Source analysis
When it's finished, you have now a new head model for this subject in your database and can be used for the next steps/(back to tutorial 20 : Head modeling)
DUNEuro options: Basic
When assuming isotropic conductivities for all the tissues, the DUNEuro basic options options are limited to the selection of the tissues and their conductivities.
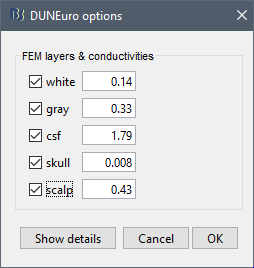
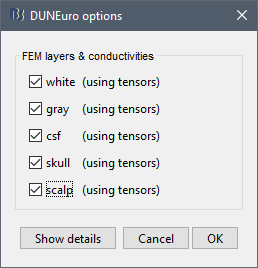
FEM layers: Brainstorm reads the lists of tissues from the selected FEM mesh. Users can select the desired layers to include in the FEM computation. According to the modalities, the recommended selections are:
EEG: Select all the layers
MEG: Select only the inner layers (inside the inner skull = brain = white+gray+csf)
SEEG: Select only the inner layers
ECOG: Select only the inner layers
- Any combination of modalities that includes MEG: select all the layers
FEM conductivities (isotropic): Left figure - Brainstorm proposes default conductivity values for each layer (derived from the SimBio software, see this article). However, you have the possibility to change these values according to your model.
FEM conductivities (anisotropic): Right figure - Advanced users can use information from diffusion imaging exams to estimate anisotropic conductivity tensors for each element of the FEM mesh. For more information, refer to the tutorial: FEM tensors estimation. To remove previously computed tensors and restore the default isotropic approach, right-click on the FEM mesh > Clear FEM tensors.
DUNEuro options: Advanced [TODO]
In the DUNEuro options panel, click on the button Show details to access the advanced options.
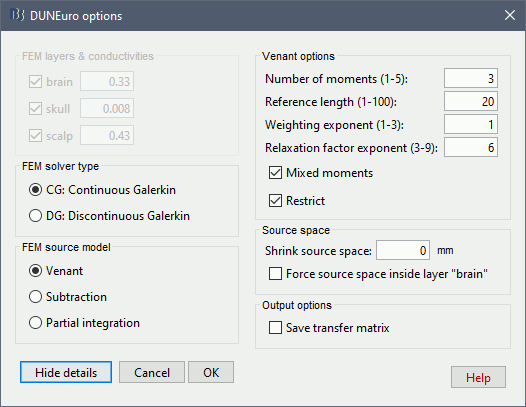
FEM layers & conductivities: Discussed in the previous section.
FEM solver type: (only the fitted FEM are integrated in Brainstorm at the moment)
CG: Continuous Galerkin: This is the standard Lagrangian method.
DC: Discontinuous Galerkin:
FEM source model:
Venant:
- Number of moments:
- Reference length:
- Weighting exponent:
- Relaxation factor exponent:
- Mixed moment:
- Restrict:
Subtraction:
- intorderadd:
- intorderadd_lb:
Partial integration:
Source space:
Shrink source space: The dipoles are moved inward by the specified distance (in mm).
Force source space: This is required when the dipoles are not within the gray matter.
Output options:
Save transfer matrix:
For more information about these methods, users can check Johannes Vorwerk's PhD thesis.
Troubleshooting [TODO]
DUNEuro binaries may crash for various reasons: we tried to list the possible causes here. Many FEM forward modeling issues are related with memory overload or extremely long computation times. Reducing the size of the problem may help in many cases. If you cannot find a solution, please post the full error message on the Brainstorm user forum (you can copy-paste the error message from the Matlab command window after closing the error message box).
Grid error messages
- Error message: Coordinate is outside of the grid, or grid is not convex
- Explanation: Some dipoles are probably outside of the cortex, users need to correct the source space.
- Solution: ?
Remove the neck
One way to reduce the size of the forward problem is to decrease the number of FEM elements in the head model. When the field of the MRI is large, you may have the mesh of the neck and even the shoulders. In most cases, it is safe to remove the lower part of the FEM mesh, below the nose and the brainstem. Right-click on the FEM mesh > Resect Neck.
Once the process is finished, a new FEM mesh appears in the database, with a tag "resect". The following figure shows the model before (743828 vertices / 4079587 elements) and after resection (613955 vertices / 3400957 elements). It will reduce the size of the problem by 20%.
Additional documentation
Related tutorials
DUNEuro references
DUNEuro wiki: https://gitlab.dune-project.org/duneuro/duneuro/wikis/home
DUNEuro website: http://duneuro.org/
List of the parameters: https://docs.google.com/spreadsheets/d/1MqURQsszn8Qj3-XRX_Z8qFFnz6Yl2-uYALkV-8pJVaM/edit#gid=0
