|
Size: 9913
Comment:
|
Size: 22764
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| == MRI visualization == * The ''Display options'' panel allows to hide some of the components. If the ''MIP/Anatomy ''checkbox is selected, the Maximum Intensity Power is displayed instead of the regular slices: At each point, the value displayed is the maximum value across all the slices. * The ''Coordinates ''panel shows the (x,y,z) coordinates in the coordinate systems managed by Brainstorm: MRI and SCS. * Once you're done, click on ''Cancel ''(unless you really want to save all your experiments). * There are many different ways to display the MR volumes. Right click on the ''T1-MRI'' in ''Default anatomy'', menu ''Display''. Just click everywhere and try all the options by yourself, it is the best way to learn.<<BR>><<BR>> {{attachment:popupMri.gif}} ==== MRI Viewer ==== Already introduced in this tutorial<<BR>>It is the default visualization mode; when you double-click on the MRI file, it brings up the MRI Viewer. ==== Axial / coronal / sagittal slices ==== {{attachment:mriSlicesContact.gif||height="231",width="210"}} |
= Tutorial 3: Display the anatomy = ''Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet'' <<TableOfContents(2,2)>> == Anatomy folder == The anatomy of the subject "Subject01" should now contain all the files Brainstorm could import from the FreeSurfer segmentation results: * '''MRI''': T1-weighted MRI, resampled and re-aligned by FreeSurfer. * '''ASEG''': Volume parcellation with the FreeSurfer subcortical regions. * '''Head mask''': Head surface, generated by Brainstorm.<<BR>>If this doesn't look good for your subject, you can recalculate another head surface using different parameters: right-click on the subject folder > [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Handling_errors|Generate head surface]]. * '''Cortex_330142V''': High-resolution pial envelope generated by FreeSurfer. * '''Cortex_15002V''': Low-resolution pial envelope, downsampled from the original one by Brainstorm. * '''White_*''': White matter envelope, high and low resolution. * '''Mid_*''': Surface that represents the mid-point between the white and cortex envelopes. * '''Subcortical''': Save FreeSurfer subcortical regions as in the ASEG volume, but tesselated as surfaces. * For more information about the files generated by FreeSurfer, read the [[Tutorials/LabelFreeSurfer|FreeSurfer page]].<<BR>><<BR>> {{attachment:import_anat_files.gif}} == Default surfaces == * There are four possible surface types: cortex, inner skull, outer skull, head. * For each type of surface, one file is selected as the one to use by default for all the operations. * This selected surface is displayed in <<HTML(<FONT color="#00DD00">)>> '''green'''<<HTML(</FONT>)>>. * Here, there is only one "head" surface, which is selected. * The mid, cortex and white surfaces can all be used as "cortex" surfaces, only one can be selected at a time. By default, the '''low-resolution cortex''' should be selected and displayed in green. * To select a different cortex surface, you can double-click on it or right-click > Set as default. == MRI Viewer == Right-click on the MRI to get the list of the available display menus: . {{attachment:mri_popup.gif}} Open the MRI Viewer. This interface was already introduced in the previous tutorial. It corresponds to the default display menu if you double-click on the MRI from the database explorer. Description of the window: * '''MIP Anatomy''': Maximum Intensity Projection. When this option is selected, the MRI viewer shows the maximum intensity value across all the slices in each direction. This maximum does not depend on the selected slice, therefore if you move the cursor, the image stays the same. * '''Neurological/Radiological''': There are two standard orientations for displaying medical scans. In the neurological orientation, the left hemisphere is on the left of the image, in the radiological orientation the left hemisphere is on the right of the image. * '''Coordinates''': Position of the cursor in different coordinate systems. See: [[CoordinateSystems]] * '''Colormap''': Click on the colorbar and move up/down (brightness) or left/right (contrast) * '''Popup menu''': All the figures have additional options available in a popup menu, accessible with a right-click on the figure. The colormap options will be described later in the tutorials, you can test the other options by yourself.<<BR>><<BR>> {{attachment:mri_viewer.gif||width="295",height="266"}} {{attachment:mri_viewer_mip.gif||width="295",height="266"}} == MRI contact sheets == You can get collections of slices in any direction (axial, coronal or sagittal) with the popup menus in the database explorer or the MRI Viewer figure. |
| Line 21: | Line 45: |
| * '''Adjust contrast''': right click + move up/down * Colormap selection in menu * Save as image in menu ==== 3D orthogonal slices ==== ||<tablewidth="200px"> {{attachment:mriSlices3d.gif||height="229",width="292"}} || {{attachment:menuShortcut.gif}} || * Simple mouse operations * '''Rotate''': click + move * '''Zoom''': mouse wheel (two finger-move on a MacBook pad) * '''Move object''': left+right click + move * '''Move MRI slices''': right click + move along direction of the slice<<BR>>(or use the ''Resect ''panel in the ''Surface ''tab) * '''Colormap contrast/brightness''': click on the colorbar, and move up/down (brightness) or left/right (contrast) * '''Reset view''': double click * '''Reset colormap''': double-click on the colorbar * Popup operations (right-click on the figure) * '''Colormap''': Full ''Anatomy ''colormap edition (detailed in next tutorial) * '''MRI Display''': For now, contains only the MIP option (Maximum Intensity Power). If checked, for each orientation: displays the maximum along all the slices instead of the proper slice. * '''Get coordinates''': Pick a point in any 3D view and get its coordinates * '''Snapshots''': save images or movies from this figure * '''Figure''': change background color, display axes and menus for an advanced figure editing and management using Matlab tools. * '''Views''': configure camera position with predefined settings. |
* '''Adjust contrast''': right click + move up/down<<BR>><<BR>> {{attachment:mri_axial.gif||width="217",height="254"}} == MRI in 3D == Right-click on the MRI file in the database explorer > Display > '''3D orthogonal slices'''. . {{attachment:mri_3d.gif||width="460",height="213"}} * Simple mouse operations:<<BR>> * '''Rotate''': Click + move. Note that two different types of rotations are available: at the center of the figure the object will follow you mouse, on the sides it will do a 2D rotation of the image. * '''Zoom''': Mouse wheel, or two finger-move on a MacBook pad. * '''Move''': Left+right click + move (or middle-click + move). * '''Colormap''': Click on the colorbar and move up/down (brightness) or left/right (contrast). * '''Reset view''': Double click anywhere on the figure. * '''Reset colormap''': Double-click on the colorbar. * '''Move slices''': Right click + move along the MRI slice axis.<<BR>>(or use the ''Resect ''panel in the ''Surface ''tab) * Popup operations (right-click on the figure): * '''Colormap''': Edit the colormap, detailed in another tutorial. * '''MRI display''': For now, contains mostly the MIP option (Maximum Intensity Projection). * '''Get coordinates''': Pick a point in any 3D view and get its coordinates. * '''Snapshots''': Save images or movies from this figure. * '''Figure''': Change some of the figure options or edit it using the Matlab tools. * '''Views''': Set one of the predefined orientation. * Note the indications in the right part of the popup menu, they represent the keyboard shortcut for each menu. |
| Line 45: | Line 69: |
| * Notice the indications in the right part of the popup menu (''CTRL+A'' in front of ''Figures>Axes'', ''"1"'' in front of ''Views>Left''), they represent the keyboard shortcut for each menu. * '''Views shortcuts (0,1,2...9 and "=")''': Remember them, they will be very useful when exploring the cortical sources. It is much faster to press a key to switch from left to right hemisphere, than having to rotate the brain with the mouse. * Surfaces tab (in the Brainstorm main window): * This panel is primarily dedicated to the surfaces display, but some controls can also be useful for the 3D MRI view. * ''Transparency ''slider * ''Smooth ''slider: it changes the threshold applied to the MRI slices. If you set it zero, you will see the full slices, as extracted from the volume. * ''Resect ''panel: you can change the position of the slices with the three sliders. == Surfaces visualization == There is only one way to display the surfaces: in 3D figures. To display a surface you can either double-click on it or right-click > Display. * The mouse and keyboard operations are the same described for the 3D MRI display also apply here. * If you display two surfaces from the same subject, they will be displayed on the same figure. * Surfaces tab: more options are available<<BR>><<BR>> {{attachment:panelSurfaces.gif}} * Open the ''Cortex ''surface and try all the buttons and sliders. Then do the same with the ''Scalp ''surface (''Head''). All the controls should have some effect on the display, except the sliders in the "''Data options''" panel. They are useful only when some additional data is mapped on the surfaces. * Multiple surfaces: If you have more than one surface in a figure, you need to select the surface you want to edit before changing its properties. The list of the available surfaces is displayed on top of the ''Surfaces ''tab. * You can also quickly add or remove surfaces in any 3D figure with the top-right buttons of the ''Surfaces ''tab. == Coordinates tab == |
* '''Views shortcuts (0,1,2...9 and [=])''': Remember them, they will be very useful when exploring the cortical sources. To switch from left to right, it is much faster to press a key than having to rotate the brain with the mouse. * '''Zoom''': Keys [+] and [-] for zooming in and out. * '''Move slices''': [x]=Sagittal, [y]=Coronal, [z]=Axial, hold [shift] for reverse direction. * Surface tab (in the main Brainstorm window, right of the database explorer): * This panel is primarily dedicated to the display of the surfaces, but some controls can also be useful for the 3D MRI view. * '''Transparency''': Changes the transparency of the slices. * '''Smooth''': Changes the background threshold applied to the MRI slices. If you set it zero, you will see the full slices, as extracted from the volume. * '''Resect''': Changes the position of the slices in the three directions. == Surfaces == To display a surface you can either double-click on it or right-click > Display. The tab "Surface" contains buttons and sliders to control the display of the surfaces. * The mouse and keyboard operations described for the 3D MRI view also apply here. * '''Smooth''': Inflates the surface to make all the parts of the cortex envelope visible. <<BR>>This is just a display option, it does not actually modify the surface. * '''Color''': Changes the color of the surface. * '''Sulci''': Shows the bottom of the cortical folds with a darker color. We recommend to keep this option selected for the cortex, it helps for the interpretation of source locations on smoothed brains. * '''Edge''': Display the faces of the surface tesselation. * '''Resect''': The sliders and the buttons Left/Right/Struct at the bottom of the panel allow you to cut the surface or reorganize the anatomical structures in various ways. * '''Multiple surfaces''': If you open two surfaces from the same subject, they will be displayed on the same figure. Then you need to select the surface you want to edit before changing its properties. The list of available surfaces is displayed at the top of the ''Surface ''tab. * At the bottom of the Surface tab, you can read the number of vertices and faces in the tesselation.<<BR>><<BR>> {{attachment:surface_options.gif||width="320",height="210"}} {{attachment:surface_mesh.gif||width="286",height="210"}} == Get coordinates == |
| Line 65: | Line 92: |
| * Right-click on the 3D figure, select "Get coordinates". A new window appears. * Click anywhere on the cortex surface: a big yellow cross appears, and the coordinates of the point are displayed in all the available coordinates systems (see page: CoordinateSystems) * MRI: MR volume, in millimeters (multiplied with the Voxsize field in the MRI file). * SCS: Subject Coordinates System, in millimeters * MNI: Montreal Neurological Institute standard coordinates * You can click on "View / MRI" to see where this point is located in the MRI, using the MRI Viewer. <<BR>><<BR>> {{attachment:panelCoord.gif}} {{attachment:panelCoordPt.gif}} == Create a subject == If you explored well, you should have found the "''New subject''" menu in the protocol's popup menu. ||<tablewidth="200px"#ffffff style="vertical-align:top; "> {{attachment:menuNewSubject.gif}} || {{attachment:createNewSubject.gif}} || * Usually, the only thing you need to do here is to edit the name of the subject. * But you can also override here the protocol's defaults regarding the sharing of anatomy and channel file between subjects and conditions. For the example, leave selected the "''Yes, use default anatomy''" option. Then click on ''Save''. * You should see a the new subject in both ''Anatomy ''and ''Functional data'' views. * In the Anatomy view, the subject node only contains a link called (Default anatomy). It is here just to remind that the subject is using the default ''MNI Colin27'' anatomy.<<BR>><<BR>> {{attachment:subjectDefaultAnat.gif}} == Change the default anatomy == When you create a new protocol, the program makes a copy of the Colin27 anatomy and sets it as the default for the protocol. It means that you will be able to use the Colin27 brain as a substitute for the subjects without an individual MRI, or as the common brain for group analysis. Other sets of MRI+surfaces are available to replace the Colin27 anatomy. Right-click on ''(Default anatomy)'' > Use template. If a package is not currently available on your system, it will be downloaded from the Brainstorm website and saved in $HOME/.brainstorm/templates. The available options are: * '''Colin27''': Average of 27 scans of the same head, processed with FreeSurfer 5.3: [[http://www.bic.mni.mcgill.ca/ServicesAtlases/Colin27|more information]] * '''Colin27_2012''': Previous version of the default anatomy distributed with Brainstorm * '''ICBM152''': Non-linear average of 152 subjects, processed with FreeSurfer 5.3: [[http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009|more information]] * '''FSAverage''': Average of 40 subjects using a spherical averaging described in [[http://nmr.mgh.harvard.edu/~fischl/reprints/morphing_human_brain_mapping_reprint.pdf|(Fischl et al. 1999)]].<<BR>>It is the default FreeSurfer brain: please [[https://surfer.nmr.mgh.harvard.edu/registration.html|register here]] if you are using it. * '''Infant7w''': 7-week infant brain with the antomical atlas presented in [[http://www.sciencedirect.com/science/article/pii/S105381191400411X|(Kabdebon et al. 2014)]]. They all include the following information: * T1 MRI volume * Cortex surface: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices) * Head surface: based on the head used for FSAverage in the MNE software * FreeSurfer spherical registration of each hemisphere, with which we can co-register the individual brains processed with FreeSurfer with the selected default anatomy * FreeSurfer surface-based atlases: Desikan-Killiany, Destrieux, Brodman, Mindboggle<<BR>>(plus Yeo2011 and PALS for FSAverage only) The atlases will be discussed in the following tutorials. For more information on the interactions between FreeSurfer and Brainstorm: [[Tutorials/LabelFreeSurfer|read this tutorial]]. {{attachment:changeDefault.gif|changeDefault1.gif}} If you click on any of the download options, it downloads it into your $HOME/.brainstorm/templates folder: {{attachment:changeDefault2.gif}} Then the list of files in the (default anatomy) folder is replaced with the new template. {{attachment:changeDefault3.gif}} == Modify the default MRI fiducials == The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm (Colin27, ICBM152, FSAverage). By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot in the [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems|Coordinates systems page]]. If you want to use an anatomy template but you are using a different convention when digitizing the position of those points, you have to modify the default positions of the template with the MRI Viewer. * Go to the anatomy view * In (default anatomy), right-click on the MRI > Edit MRI * Modify the position of the fiducial points to match your own convention * Click on [Save], it will update the surfaces to match the new coordinate system |
* Right-click on the 3D figure, select "Get coordinates". * Click anywhere on the cortex surface: a yellow cross appears and the coordinates of the point are displayed in all the available coordinates systems. * You can click on [View/MRI] to see where this point is located in the MRI, using the MRI Viewer. <<BR>><<BR>> {{attachment:surface_coordinates.gif||width="365",height="152"}} == Subcortical regions: Volume == The standard FreeSurfer segmentation pipeline generates multiple volume parcellations of anatomical regions. This example dataset includes only the ASEG parcellation, documenting the subcortical structures. Double-click on the volume parcellation '''ASEG''' to open it for display. This would open the MRI Viewer with two volumes: the T1 MRI as the background, and the ASEG volume as a semi-transparent overlay. * Adjust the transparency of the ASEG overlay from the Surface tab, slider Transp. * The '''name of the region''' under the cursor appears at the top-right corner. The integer before this name is the label of the ROI, ie. the integer value of the voxel under the cursor in the ASEG volume.<<BR>><<BR>> {{attachment:aseg_volume.gif}} * Close the MRI viewer. * Double-click again on the subject's MRI to open it in the MRI viewer. * Observe that the anatomical label is also present at the top-right corner of this figure; in this case, the integer reprents the voxel value of the displayed MRI. This label information comes from the ASEG file: whenever there are volume parcellations available for the subject, one of them is loaded in the MRI Viewer by default. The name of the selected parcellation is displayed in the figure title bar. * You can change the selected parcellation with the right-click popup menu '''Anatomical atlas'''. You can change the parcellation scheme, disable its use to make the MRI work faster, or show the parcellation volume as an overlay (menu Show atlas). <<BR>><<BR>> {{attachment:aseg_label.gif}} == Subcortical regions: Surface == Brainstorm reads the ASEG volume labels and tesselates some of these regions, then groups all the meshes in a large surface file where the regions are identified in an atlas called "Structures". It identifies: 8 bilateral structures (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, cerebellum) and 1 central structure (brainstem). These structures can be useful for advanced source modeling, but will not be used in the introduction tutorials. Please refer to the advanced tutorials for more information: [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_scouts|Volume source estimation]] and [[http://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas#Source_estimation|Deep cerebral structures]]. . {{attachment:aseg.gif||width="368",height="186"}} With the button '''[Struct]''' at the bottom of the Surface tab, you can see the structures separately. . {{attachment:resect_struct.gif||width="432",height="226"}} == Registration MRI/surfaces == The MRI and the surfaces are represented using the different coordinate systems and could be misregistered for various reasons. If you are using the automated segmentation pipeline from FreeSurfer or BrainSuite you should never have any problem, but if something goes wrong or in the case of more manual import procedures it is always good to check that the MRI and the surfaces are correctly aligned. * Right-click on the low-res cortex > MRI Registration > Check MRI/surface registration * The calculation of the interpolation between the MRI and the cortex surface takes a few seconds, but the result is then saved in the database and will be reused later. * The yellow lines represent the re-interpolation of the surface in the MRI volume.<<BR>><<BR>> {{attachment:surface_register.gif||width="271",height="216"}} <<TAG(Advanced)>> == Interaction with the file system == For most manipulations, it is not necessary to know exactly what is going on at the level of the file system, in the Brainstorm database directory. However, many things are not accessible from the Brainstorm interface, you may sometimes find it useful to manipulate some piece of data directly from the Matlab command window. ==== Where are the files ? ==== * Leave your mouse for a few seconds over any node in the database explorer, a tooltip will appear with the name and path of the corresponding file on the hard drive. * Paths are relative to current protocol path (brainstorm_db/TutorialIntroduction). What is displayed in the Brainstorm window is a comment and may have nothing to do with the real file name. For instance, the file name corresponding to "head mask" is ''Subjec01/tess_head_mask.mat''. * Almost all the files in the database are in Matlab .mat format. You can load and edit them easily in the Matlab environment, where they appear as structures with several fields.<<BR>><<BR>> {{attachment:file_tooltip.gif||width="482",height="122"}} ==== Popup menu: File ==== Right-click on a surface file: many menus can lead you to the files and their contents. . {{attachment:file_menu.gif}} * '''View file contents''': Display all the fields in the Matlab .mat file. <<BR>><<BR>> {{attachment:file_contents.gif}} * '''View file history''': Review the History field in the file, that records all the operations that were performed on the file since if was imported in Brainstorm.<<BR>><<BR>> {{attachment:file_history.gif}} * '''Export to file''': Export in one of the supported mesh file format. * '''Export to Matlab''': Load the contents of the .mat file in the Matlab base workspace. It is then accessible from the Matlab command window. * '''Import from Matlab''': Replace the selected file with the content of a variable from the Matlab base workspace. Useful to save back in the database a structure that was exported and modified manually with the Matlab command window. * '''Copy / Cut / Paste''': Allow you to copy/move files in the database explorer. Keyboard shortcuts for these menus are the standard Windows shortcuts (Ctrl+C, Ctrl+X, Ctrl+V). The database explorer also supports drag-and-drop operations for moving files between different folders. * '''Delete''': Delete a file. Keyboard shortcuts: Delete key. * '''Rename''': Change the Comment field in the file. It "renames" the file in the database explorer, but does not change the actual file name on the hard drive. Keyboard shortcut: F2 * '''Copy file path to clipboard''': Copies the full file name into the system clipboard, so that you can paste it in any other window (Ctrl+V or Paste menu) * '''Go to this directory (Matlab)''': Change the current Matlab path, so that you can access the file from the Matlab ''Command window'' or the Matlab ''Current directory'' window * '''Show in file explorer''': Open a file explorer window in this directory. * '''Open terminal in this folder''': Start a system console in the file directory (Linux and MacOS only). ==== What are all these other files ? ==== * If you look in ''brainstorm_db/TutorialIntroduction ''with the file explorer of your operating system, you'll find many other directories and files that are not visible in the database explorer.<<BR>><<BR>> {{attachment:file_disk.gif}} * The protocol TutorialIntroduction is divided in ''Anatomy ''and ''Datasets ''directories: * Each subject in ''anat ''is described by an extra file: ''brainstormsubject.mat'' * Each folder in ''data ''is described by an extra file: ''brainstormstudy.mat'' * '''anat/@default_subject''': Contains the files of the default anatomy ''(Default anatomy)'' * '''data/@default_study''': Files shared between different subjects ''(Global common files)'' * '''data/@inter''': Results of inter-subject analysis * '''data/Subject01/@default_study''': Files shared between different folders in Subject01 * '''data/Subject01/@intra''': Results of intra-subject analysis (across different folders) <<TAG(Advanced)>> == On the hard drive: MRI == Right-click on the MRI > File > View file contents: . {{attachment:contents_mri.gif||width="530"}} ==== Structure of the MRI files: subjectimage_*.mat ==== * '''Comment''': String displayed in the database explorer to represent the file. * '''Cube''': [Nsagittal x Ncoronal x Naxial] full MRI volume. * '''Voxsize''': Size of one voxel in millimeters (sagittal, coronal, axial). * '''SCS''': Defines the [[CoordinateSystems|Subject Coordinate System]]. Points below are in MRI (millimeters) coordinates. * '''NAS''': (x,y,z) coordinates of the nasion fiducial. * '''LPA''': (x,y,z) coordinates of the left ear fiducial. * '''RPA''': (x,y,z) coordinates of the right ear fiducial. * '''R''': [3x3] rotation matrix from MRI coordinates to SCS coordinates. * '''T''': [3x1] translation matrix from MRI coordinates to SCS coordinates. * '''Origin''': MRI coordinates of the point with SCS coordinates (0,0,0). * '''NCS''': Defines the [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|MNI coordinate system]], either with a linear or a non-linear transformation. * '''AC''': (x,y,z) coordinates of the Anterior Commissure. * '''PC''': (x,y,z) coordinates of the Posterior Commissure. * '''IH''': (x,y,z) coordinates of an Inter-Hemisperic point. * (Linear transformation) * '''R''': [3x3] rotation matrix from MRI coordinates to MNI coordinates. * '''T''': [3x1] translation matrix from MRI coordinates to MNI coordinates. * (Non-linear transformation) * '''iy''': 3D floating point matrix: Inverse MNI deformation field, as in SPM naming conventions. Same size as the Cube matrix, it gives for each voxel its coordinates in the MNI space, and is therefore used to convert from MRI coordinates to MNI coordinates. * '''y''': 3D floating point matrix: Forward MNI deformation field, as in SPM naming conventions. For some MNI coordinates, it gives their coorrespondance in the original MRI space. To be interpreted, it has to be used with the matrix y_vox2ras. * '''y_vox2ras''': [4x4 double], transformation matrix that converts from voxel coordinates of the y volume to MNI coordinates. * '''y_method''': Algorithm used for computing the normalization ('segment'=SPM12 Segment) * '''Origin''': MRI coordinates of the point with NCS coordinates (0,0,0). * '''Header''': Header from the original file format (.nii, .mgz, ...) * '''Histogram''': Result of the internal analysis of the MRI histogram, mainly to detect background level. * '''InitTransf''': [Ntransform x 2] cell-matrix: Transformations that are applied to the MRI before importing the surfaces. Example: {'vox2ras', [4x4 double]} * '''Labels''': [Nlabels x 3] cell-matrix: For anatomical parcellations, this field contains the names and RGB colors associated with each integer label in the volume. Example:<<BR>>{0, 'Background', [0 0 0]}<<BR>>{1, 'Precentral L', [203 142 203]} * '''History''': List of operations performed on this file (menu File > View file history). ==== Useful functions ==== * /toolbox/io/'''in_mri_bst'''(MriFile): Read a Brainstorm MRI file and compute the missing fields. * /toolbox/io/'''in_mri'''(MriFile, FileFormat=[]): Read a MRI file (format is auto-detected). * /toolbox/io/'''in_mri_*.m''': Low-level functions for reading all the file formats. * /toolbox/anatomy/'''mri_*.m''': Routines for manipulating MRI volumes. * /toolbox/gui/'''view_mri'''(MriFile, ...): Display an imported MRI in the MRI viewer. * /toolbox/gui/'''view_mri_3d'''(MriFile, ...): Display an imported MRI in a 3D figure. <<TAG(Advanced)>> == On the hard drive: Surface == Right-click on any cortex surface > File > View file contents: . {{attachment:file_contents.gif||width="529",height="271"}} ==== Structure of the surface files: tess_*.mat ==== * '''Atlas''': Array of structures, each entry is one menu in the drop-down list in the Scout tab. * '''Name''': Label of the atlas (reserved names: "User scouts", "Structures", "Source model") * '''Scouts''': List of regions of interest in this atlas, see the [[http://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#On_the_hard_drive|Scout tutorial]]. * '''Comment''': String displayed in the database explorer to represent the file. '' '' * '''Curvature''': [Nvertices x 1], curvature value at each point. * '''Faces''': [Nfaces x 3], triangles constituting the surface mesh. * '''History''': List of operations performed on this file (menu File > View file history). * '''iAtlas''': Index of the atlas that is currently selected for this surface. * '''Reg''': Structure with registration information, used to interpolate the subject's maps on a template. * '''Sphere.Vertices''': Location of the surface vertices on the [[Tutorials/CoregisterSubjects|FreeSurfer registered spheres]]. * '''Square.Vertices''': Location of the surface vertices in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects#Registration_with_BrainSuite|BrainSuite atlas]]. * '''AtlasSquare.Vertices''': Corresponding vertices in the high-resolution [[http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects#Registration_with_BrainSuite|BrainSuite atlas]]. * '''SulciMap''': [Nvertices x 1], binary mask marking the botton of the sulci (1=displayed as darker). * '''tess2mri_interp''': [Nvoxels x Nvertices] sparse interpolation matrix MRI<=>surface. * '''VertConn''': [Nvertices x Nvertices] Sparse adjacency matrix, VertConn(i,j)=1 if i and j are neighbors. * '''Vertices''': [Nvertices x 3], coordinates (x,y,z) of all the points of the surface, in SCS coordinates. * '''VertNormals''': [Nvertices x 3], direction (x,y,z) of the normal to the surface at each vertex. ==== Useful functions ==== * /toolbox/io/'''in_tess_bst'''(SurfaceFile): Read a Brainstorm surface file and compute the missing fields. * /toolbox/io/'''in_tess'''(TessFile, FileFormat=[], sMri=[]): Read a surface file (format is auto-detected). * /toolbox/io/'''in_tess_*.m''': Low-level functions for reading all the file formats. * /toolbox/anatomy/'''tess_*.m''': Routines for manipulating surfaces. * /toolbox/gui/'''view_surface'''(SurfaceFile, ...): Display an imported surface in a 3D figure. * /toolbox/gui/'''view_surface_data'''(SurfaceFile, OverlayFile, ...): Display a surface with a source map. * /toolbox/gui/'''view_surface_matrix'''(Vertices, Faces, ...): Display a mesh in a 3D figure. <<HTML(<!-- END-PAGE -->)>> <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials/ImportAnatomy&next=Tutorials/ChannelFile")>> <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/ExploreAnatomy)>> |
Tutorial 3: Display the anatomy
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
Contents
Anatomy folder
The anatomy of the subject "Subject01" should now contain all the files Brainstorm could import from the FreeSurfer segmentation results:
MRI: T1-weighted MRI, resampled and re-aligned by FreeSurfer.
ASEG: Volume parcellation with the FreeSurfer subcortical regions.
Head mask: Head surface, generated by Brainstorm.
If this doesn't look good for your subject, you can recalculate another head surface using different parameters: right-click on the subject folder > Generate head surface.Cortex_330142V: High-resolution pial envelope generated by FreeSurfer.
Cortex_15002V: Low-resolution pial envelope, downsampled from the original one by Brainstorm.
White_*: White matter envelope, high and low resolution.
Mid_*: Surface that represents the mid-point between the white and cortex envelopes.
Subcortical: Save FreeSurfer subcortical regions as in the ASEG volume, but tesselated as surfaces.
For more information about the files generated by FreeSurfer, read the FreeSurfer page.
Default surfaces
- There are four possible surface types: cortex, inner skull, outer skull, head.
- For each type of surface, one file is selected as the one to use by default for all the operations.
This selected surface is displayed in green.
- Here, there is only one "head" surface, which is selected.
The mid, cortex and white surfaces can all be used as "cortex" surfaces, only one can be selected at a time. By default, the low-resolution cortex should be selected and displayed in green.
To select a different cortex surface, you can double-click on it or right-click > Set as default.
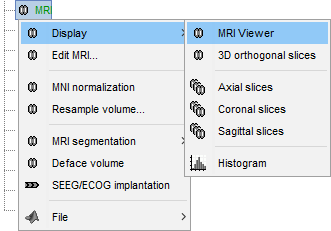
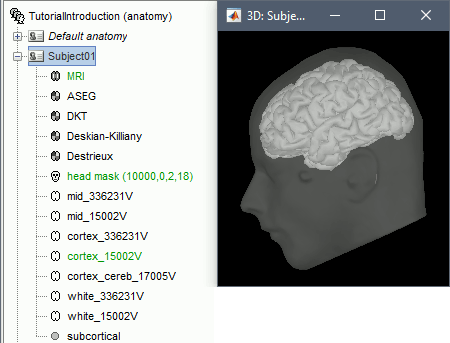
MRI Viewer
Right-click on the MRI to get the list of the available display menus:
Open the MRI Viewer. This interface was already introduced in the previous tutorial. It corresponds to the default display menu if you double-click on the MRI from the database explorer. Description of the window:
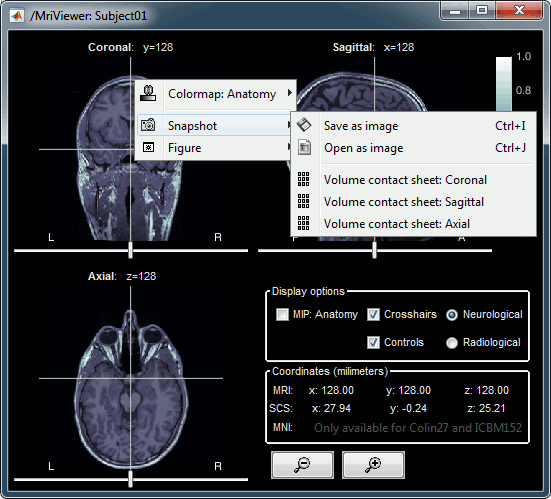
MIP Anatomy: Maximum Intensity Projection. When this option is selected, the MRI viewer shows the maximum intensity value across all the slices in each direction. This maximum does not depend on the selected slice, therefore if you move the cursor, the image stays the same.
Neurological/Radiological: There are two standard orientations for displaying medical scans. In the neurological orientation, the left hemisphere is on the left of the image, in the radiological orientation the left hemisphere is on the right of the image.
Coordinates: Position of the cursor in different coordinate systems. See: CoordinateSystems
Colormap: Click on the colorbar and move up/down (brightness) or left/right (contrast)
Popup menu: All the figures have additional options available in a popup menu, accessible with a right-click on the figure. The colormap options will be described later in the tutorials, you can test the other options by yourself.
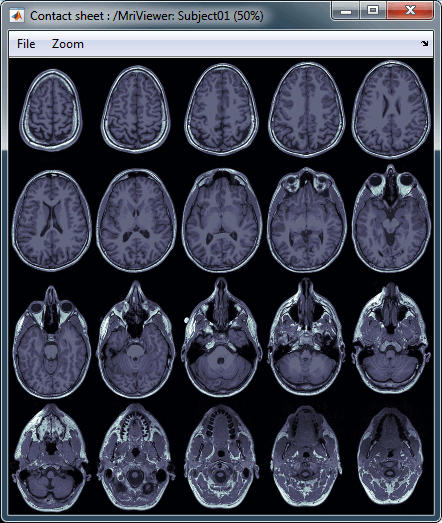
MRI contact sheets
You can get collections of slices in any direction (axial, coronal or sagittal) with the popup menus in the database explorer or the MRI Viewer figure.
Zoom: mouse wheel (or two finger-move on a MacBook pad)
Move in zoomed image: click + move
Adjust contrast: right click + move up/down
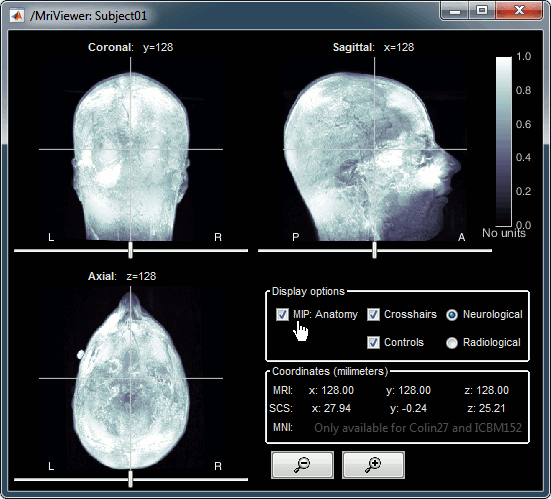
MRI in 3D
Right-click on the MRI file in the database explorer > Display > 3D orthogonal slices.
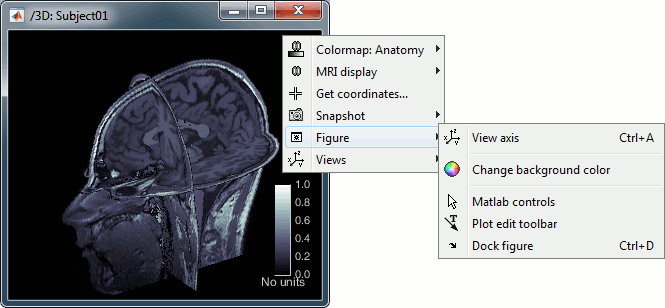
Simple mouse operations:
Rotate: Click + move. Note that two different types of rotations are available: at the center of the figure the object will follow you mouse, on the sides it will do a 2D rotation of the image.
Zoom: Mouse wheel, or two finger-move on a MacBook pad.
Move: Left+right click + move (or middle-click + move).
Colormap: Click on the colorbar and move up/down (brightness) or left/right (contrast).
Reset view: Double click anywhere on the figure.
Reset colormap: Double-click on the colorbar.
Move slices: Right click + move along the MRI slice axis.
(or use the Resect panel in the Surface tab)
- Popup operations (right-click on the figure):
Colormap: Edit the colormap, detailed in another tutorial.
MRI display: For now, contains mostly the MIP option (Maximum Intensity Projection).
Get coordinates: Pick a point in any 3D view and get its coordinates.
Snapshots: Save images or movies from this figure.
Figure: Change some of the figure options or edit it using the Matlab tools.
Views: Set one of the predefined orientation.
- Note the indications in the right part of the popup menu, they represent the keyboard shortcut for each menu.
- Keyboard shortcuts:
Views shortcuts (0,1,2...9 and [=]): Remember them, they will be very useful when exploring the cortical sources. To switch from left to right, it is much faster to press a key than having to rotate the brain with the mouse.
Zoom: Keys [+] and [-] for zooming in and out.
Move slices: [x]=Sagittal, [y]=Coronal, [z]=Axial, hold [shift] for reverse direction.
- Surface tab (in the main Brainstorm window, right of the database explorer):
- This panel is primarily dedicated to the display of the surfaces, but some controls can also be useful for the 3D MRI view.
Transparency: Changes the transparency of the slices.
Smooth: Changes the background threshold applied to the MRI slices. If you set it zero, you will see the full slices, as extracted from the volume.
Resect: Changes the position of the slices in the three directions.
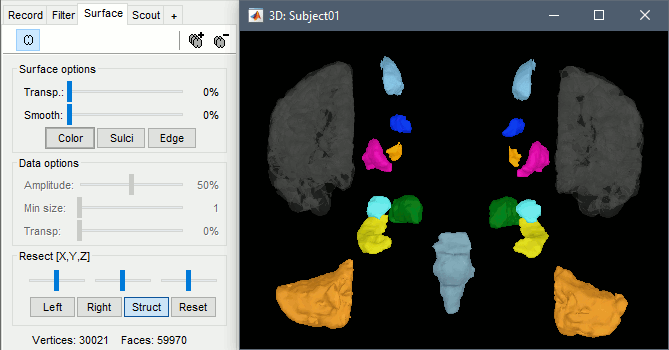
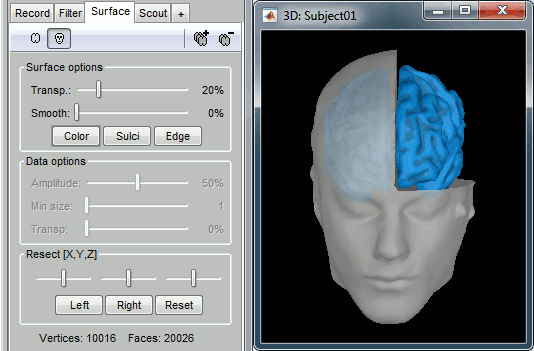
Surfaces
To display a surface you can either double-click on it or right-click > Display. The tab "Surface" contains buttons and sliders to control the display of the surfaces.
- The mouse and keyboard operations described for the 3D MRI view also apply here.
Smooth: Inflates the surface to make all the parts of the cortex envelope visible.
This is just a display option, it does not actually modify the surface.Color: Changes the color of the surface.
Sulci: Shows the bottom of the cortical folds with a darker color. We recommend to keep this option selected for the cortex, it helps for the interpretation of source locations on smoothed brains.
Edge: Display the faces of the surface tesselation.
Resect: The sliders and the buttons Left/Right/Struct at the bottom of the panel allow you to cut the surface or reorganize the anatomical structures in various ways.
Multiple surfaces: If you open two surfaces from the same subject, they will be displayed on the same figure. Then you need to select the surface you want to edit before changing its properties. The list of available surfaces is displayed at the top of the Surface tab.
At the bottom of the Surface tab, you can read the number of vertices and faces in the tesselation.
Get coordinates
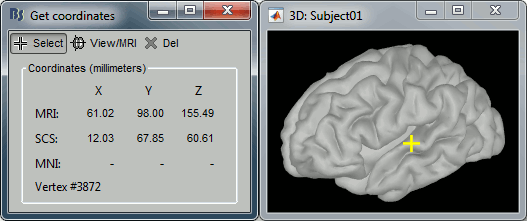
- Close all the figures. Open the cortex surface again.
- Right-click on the 3D figure, select "Get coordinates".
- Click anywhere on the cortex surface: a yellow cross appears and the coordinates of the point are displayed in all the available coordinates systems.
You can click on [View/MRI] to see where this point is located in the MRI, using the MRI Viewer.
Subcortical regions: Volume
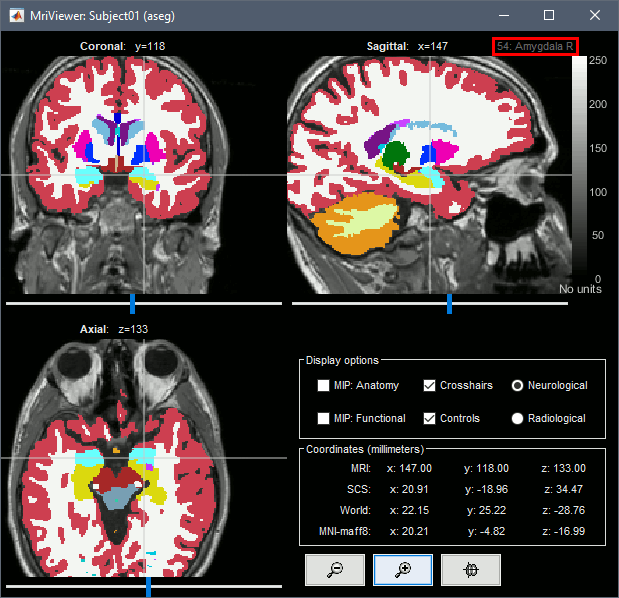
The standard FreeSurfer segmentation pipeline generates multiple volume parcellations of anatomical regions. This example dataset includes only the ASEG parcellation, documenting the subcortical structures. Double-click on the volume parcellation ASEG to open it for display. This would open the MRI Viewer with two volumes: the T1 MRI as the background, and the ASEG volume as a semi-transparent overlay.
- Adjust the transparency of the ASEG overlay from the Surface tab, slider Transp.
The name of the region under the cursor appears at the top-right corner. The integer before this name is the label of the ROI, ie. the integer value of the voxel under the cursor in the ASEG volume.
- Close the MRI viewer.
- Double-click again on the subject's MRI to open it in the MRI viewer.
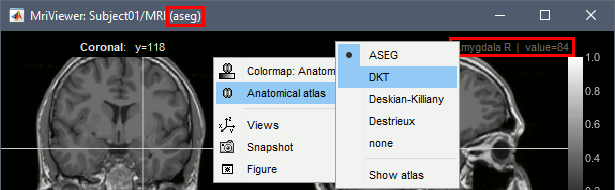
- Observe that the anatomical label is also present at the top-right corner of this figure; in this case, the integer reprents the voxel value of the displayed MRI. This label information comes from the ASEG file: whenever there are volume parcellations available for the subject, one of them is loaded in the MRI Viewer by default. The name of the selected parcellation is displayed in the figure title bar.
You can change the selected parcellation with the right-click popup menu Anatomical atlas. You can change the parcellation scheme, disable its use to make the MRI work faster, or show the parcellation volume as an overlay (menu Show atlas).
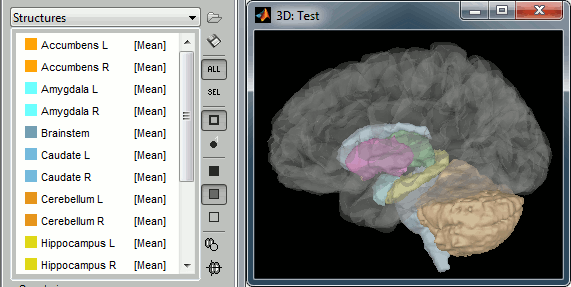
Subcortical regions: Surface
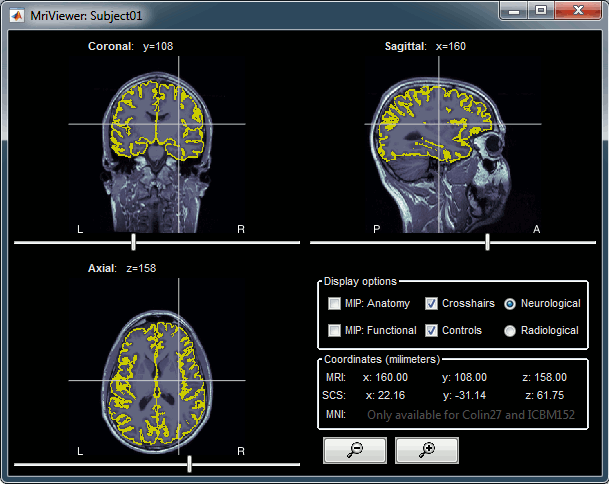
Brainstorm reads the ASEG volume labels and tesselates some of these regions, then groups all the meshes in a large surface file where the regions are identified in an atlas called "Structures". It identifies: 8 bilateral structures (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, cerebellum) and 1 central structure (brainstem).
These structures can be useful for advanced source modeling, but will not be used in the introduction tutorials. Please refer to the advanced tutorials for more information: Volume source estimation and Deep cerebral structures.
With the button [Struct] at the bottom of the Surface tab, you can see the structures separately.
Registration MRI/surfaces
The MRI and the surfaces are represented using the different coordinate systems and could be misregistered for various reasons. If you are using the automated segmentation pipeline from FreeSurfer or BrainSuite you should never have any problem, but if something goes wrong or in the case of more manual import procedures it is always good to check that the MRI and the surfaces are correctly aligned.
Right-click on the low-res cortex > MRI Registration > Check MRI/surface registration
- The calculation of the interpolation between the MRI and the cortex surface takes a few seconds, but the result is then saved in the database and will be reused later.
The yellow lines represent the re-interpolation of the surface in the MRI volume.
Interaction with the file system
For most manipulations, it is not necessary to know exactly what is going on at the level of the file system, in the Brainstorm database directory. However, many things are not accessible from the Brainstorm interface, you may sometimes find it useful to manipulate some piece of data directly from the Matlab command window.
Where are the files ?
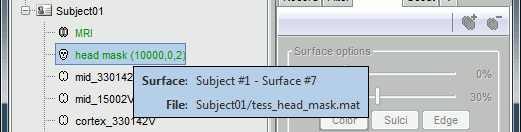
- Leave your mouse for a few seconds over any node in the database explorer, a tooltip will appear with the name and path of the corresponding file on the hard drive.
Paths are relative to current protocol path (brainstorm_db/TutorialIntroduction). What is displayed in the Brainstorm window is a comment and may have nothing to do with the real file name. For instance, the file name corresponding to "head mask" is Subjec01/tess_head_mask.mat.
Almost all the files in the database are in Matlab .mat format. You can load and edit them easily in the Matlab environment, where they appear as structures with several fields.
Popup menu: File
Right-click on a surface file: many menus can lead you to the files and their contents.
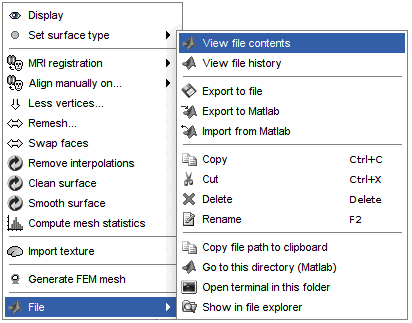
View file contents: Display all the fields in the Matlab .mat file.
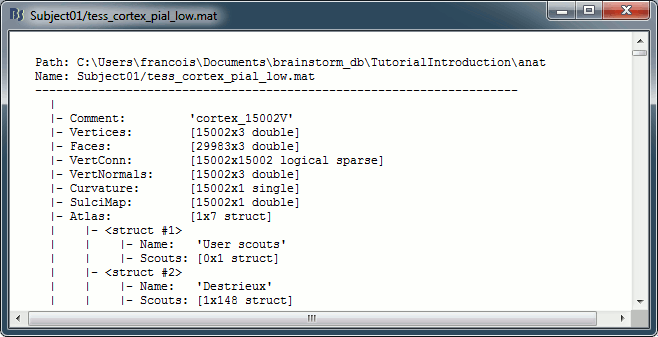
View file history: Review the History field in the file, that records all the operations that were performed on the file since if was imported in Brainstorm.
Export to file: Export in one of the supported mesh file format.
Export to Matlab: Load the contents of the .mat file in the Matlab base workspace. It is then accessible from the Matlab command window.
Import from Matlab: Replace the selected file with the content of a variable from the Matlab base workspace. Useful to save back in the database a structure that was exported and modified manually with the Matlab command window.
Copy / Cut / Paste: Allow you to copy/move files in the database explorer. Keyboard shortcuts for these menus are the standard Windows shortcuts (Ctrl+C, Ctrl+X, Ctrl+V). The database explorer also supports drag-and-drop operations for moving files between different folders.
Delete: Delete a file. Keyboard shortcuts: Delete key.
Rename: Change the Comment field in the file. It "renames" the file in the database explorer, but does not change the actual file name on the hard drive. Keyboard shortcut: F2
Copy file path to clipboard: Copies the full file name into the system clipboard, so that you can paste it in any other window (Ctrl+V or Paste menu)
Go to this directory (Matlab): Change the current Matlab path, so that you can access the file from the Matlab Command window or the Matlab Current directory window
Show in file explorer: Open a file explorer window in this directory.
Open terminal in this folder: Start a system console in the file directory (Linux and MacOS only).
What are all these other files ?
If you look in brainstorm_db/TutorialIntroduction with the file explorer of your operating system, you'll find many other directories and files that are not visible in the database explorer.
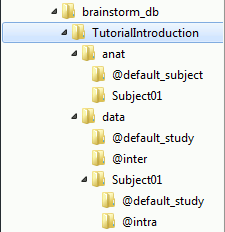
The protocol TutorialIntroduction is divided in Anatomy and Datasets directories:
Each subject in anat is described by an extra file: brainstormsubject.mat
Each folder in data is described by an extra file: brainstormstudy.mat
anat/@default_subject: Contains the files of the default anatomy (Default anatomy)
data/@default_study: Files shared between different subjects (Global common files)
data/@inter: Results of inter-subject analysis
data/Subject01/@default_study: Files shared between different folders in Subject01
data/Subject01/@intra: Results of intra-subject analysis (across different folders)
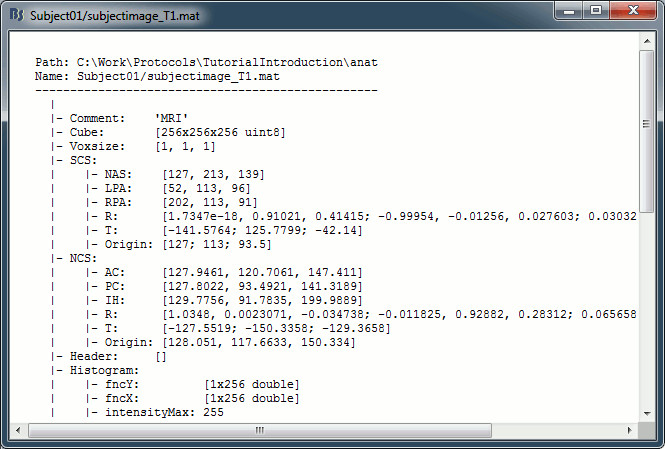
On the hard drive: MRI
Right-click on the MRI > File > View file contents:
Structure of the MRI files: subjectimage_*.mat
Comment: String displayed in the database explorer to represent the file.
Cube: [Nsagittal x Ncoronal x Naxial] full MRI volume.
Voxsize: Size of one voxel in millimeters (sagittal, coronal, axial).
SCS: Defines the Subject Coordinate System. Points below are in MRI (millimeters) coordinates.
NAS: (x,y,z) coordinates of the nasion fiducial.
LPA: (x,y,z) coordinates of the left ear fiducial.
RPA: (x,y,z) coordinates of the right ear fiducial.
R: [3x3] rotation matrix from MRI coordinates to SCS coordinates.
T: [3x1] translation matrix from MRI coordinates to SCS coordinates.
Origin: MRI coordinates of the point with SCS coordinates (0,0,0).
NCS: Defines the MNI coordinate system, either with a linear or a non-linear transformation.
AC: (x,y,z) coordinates of the Anterior Commissure.
PC: (x,y,z) coordinates of the Posterior Commissure.
IH: (x,y,z) coordinates of an Inter-Hemisperic point.
- (Linear transformation)
R: [3x3] rotation matrix from MRI coordinates to MNI coordinates.
T: [3x1] translation matrix from MRI coordinates to MNI coordinates.
- (Non-linear transformation)
iy: 3D floating point matrix: Inverse MNI deformation field, as in SPM naming conventions. Same size as the Cube matrix, it gives for each voxel its coordinates in the MNI space, and is therefore used to convert from MRI coordinates to MNI coordinates.
y: 3D floating point matrix: Forward MNI deformation field, as in SPM naming conventions. For some MNI coordinates, it gives their coorrespondance in the original MRI space. To be interpreted, it has to be used with the matrix y_vox2ras.
y_vox2ras: [4x4 double], transformation matrix that converts from voxel coordinates of the y volume to MNI coordinates.
y_method: Algorithm used for computing the normalization ('segment'=SPM12 Segment)
Origin: MRI coordinates of the point with NCS coordinates (0,0,0).
Header: Header from the original file format (.nii, .mgz, ...)
Histogram: Result of the internal analysis of the MRI histogram, mainly to detect background level.
InitTransf: [Ntransform x 2] cell-matrix: Transformations that are applied to the MRI before importing the surfaces. Example: {'vox2ras', [4x4 double]}
Labels: [Nlabels x 3] cell-matrix: For anatomical parcellations, this field contains the names and RGB colors associated with each integer label in the volume. Example:<<BR>>{0, 'Background', [0 0 0]}
{1, 'Precentral L', [203 142 203]}History: List of operations performed on this file (menu File > View file history).
Useful functions
/toolbox/io/in_mri_bst(MriFile): Read a Brainstorm MRI file and compute the missing fields.
/toolbox/io/in_mri(MriFile, FileFormat=[]): Read a MRI file (format is auto-detected).
/toolbox/io/in_mri_*.m: Low-level functions for reading all the file formats.
/toolbox/anatomy/mri_*.m: Routines for manipulating MRI volumes.
/toolbox/gui/view_mri(MriFile, ...): Display an imported MRI in the MRI viewer.
/toolbox/gui/view_mri_3d(MriFile, ...): Display an imported MRI in a 3D figure.
On the hard drive: Surface
Right-click on any cortex surface > File > View file contents:
Structure of the surface files: tess_*.mat
Atlas: Array of structures, each entry is one menu in the drop-down list in the Scout tab.
Name: Label of the atlas (reserved names: "User scouts", "Structures", "Source model")
Scouts: List of regions of interest in this atlas, see the Scout tutorial.
Comment: String displayed in the database explorer to represent the file.
Curvature: [Nvertices x 1], curvature value at each point.
Faces: [Nfaces x 3], triangles constituting the surface mesh.
History: List of operations performed on this file (menu File > View file history).
iAtlas: Index of the atlas that is currently selected for this surface.
Reg: Structure with registration information, used to interpolate the subject's maps on a template.
Sphere.Vertices: Location of the surface vertices on the FreeSurfer registered spheres.
Square.Vertices: Location of the surface vertices in the BrainSuite atlas.
AtlasSquare.Vertices: Corresponding vertices in the high-resolution BrainSuite atlas.
SulciMap: [Nvertices x 1], binary mask marking the botton of the sulci (1=displayed as darker).
tess2mri_interp: [Nvoxels x Nvertices] sparse interpolation matrix MRI<=>surface.
VertConn: [Nvertices x Nvertices] Sparse adjacency matrix, VertConn(i,j)=1 if i and j are neighbors.
Vertices: [Nvertices x 3], coordinates (x,y,z) of all the points of the surface, in SCS coordinates.
VertNormals: [Nvertices x 3], direction (x,y,z) of the normal to the surface at each vertex.
Useful functions
/toolbox/io/in_tess_bst(SurfaceFile): Read a Brainstorm surface file and compute the missing fields.
/toolbox/io/in_tess(TessFile, FileFormat=[], sMri=[]): Read a surface file (format is auto-detected).
/toolbox/io/in_tess_*.m: Low-level functions for reading all the file formats.
/toolbox/anatomy/tess_*.m: Routines for manipulating surfaces.
/toolbox/gui/view_surface(SurfaceFile, ...): Display an imported surface in a 3D figure.
/toolbox/gui/view_surface_data(SurfaceFile, OverlayFile, ...): Display a surface with a source map.
/toolbox/gui/view_surface_matrix(Vertices, Faces, ...): Display a mesh in a 3D figure.