|
Size: 6757
Comment:
|
Size: 61008
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 8: | Line 8: |
| Requirements: You need a license for the Matlab environment in order to use these tools and execute custom scripts. If you are running the compiled version of Brainstorm with the MCR library, the only custom code you can run is through the menu File > Matlab console and the process "Run Matlab command". |
|
| Line 10: | Line 12: |
| == Workflow == This section proposes a standard workflow for processing a full group study with Brainstorm. It contains the same steps of analysis as the introduction tutorials, but separating what can be done automatically from what can The list below contains all the steps of analysis that were detailed in the introduction tutorials, organized in a way that matches the actual work of the person in charge of the analyses. A good approach can be to write separate scripts, and between them perform some verifications and manual adjustments. * '''Prototype''': Start by processing one or two subjects completely interactively (exactly like in the introduction tutorials). Use the few pilot subjects that you have for your study to prototype the analysis pipeline and check manually all the intermediate stages. Take notes of what you're doing along the way, so that you can later write a script that reproduces the same operations. |
== Starting a new script == The easiest way to get started with a new Brainstorm script is to use the script generator, already introduced in the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor#Saving_a_pipeline|Select files and run processes]]. Select some files in the Process1 or Process2 tabs, select a list of processes, and use the menu '''Generate .m script'''. The example below should work on the the protocol "TutorialIntroduction" created during the introduction tutorials. * In the Process1 tab, leave the selection box empty and click on [Run]. Instead of selecting the files from the Brainstorm interface, we will select them directly from the database using a script. * Select process '''File > Select files: Recordings''' (do not execute immediately)<<BR>>Subject='''Subject01''', Condition='''[Empty]''', File comment='''Avg: deviant''' (the space is important). <<BR>><<BR>> {{attachment:start1.gif||height="334",width="400"}} * This process selects all the recordings with a comment including the string "Avg: deviant", from all the folders in Subject01 (except "Intra-subject" and "Common files"). We expect to get two files: the averages of the deviant condition for both runs. * Add process '''Pre-process > Band-pass filter''': Lower cutoff='''0Hz''', Upper cutoff='''30Hz''', Mirror. <<BR>> '''Add process File > Save snapshot''': Recordings time series, Sensors=MEG. <<BR>> <<BR>> {{attachment:start2.gif||height="294",width="518"}} * This will apply a low-pass filter at 30Hz and save a screen capture of the signals in the report. * Do not run the pipeline, select the menu '''Generate .m script '''instead. It saves a new .m file and opens it in the Matlab editor. Close the pipeline editor window and look at the script.<<BR>><<BR>> {{attachment:start3.gif||height="272",width="673"}} * The script you just generated can be the starting point to your own custom script. The following sections explain line by line how they work and how to edit them. == Line by line: Header == {{{ % Script generated by Brainstorm (19-Jul-2016) }}} All the lines starting with a "%" are comments, they are never executed. {{{ % Input files sFiles = []; SubjectNames = {... 'Subject01'}; }}} These lines define the script inputs: * '''sFiles''': The list of files in input. Currently empty because we did not select anything in input in the Process1 list. If we had selected files, it would contain a cell array of strings with relative file paths. * '''SubjectNames''': List of subject names that are used in the script. Most of the times, the generated script would contain only one entry, but it written as a cell array so that it is easier to extend it to multiple subjects with a loop (described further in this tutorial). {{{ % Start a new report bst_report('Start', sFiles); }}} Start a new report of activity: Clears all the previous logs and gets ready to record new messages. The report will collect all the messages that are generated during the execution of the script by the various processes. You can explicitely add screen captures and additional messages to the current report with the function bst_report. This report will remain open until the function bst_report('Start') is called again. To display the current report, use the menu [[http://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor#Report_viewer|File > Report viewer]]. The syntax '''function_name'''(''''SubFunction'''', arguments) is used a lot in Brainstorm, to call a subfunction available inside a .m file. This line above calls the function Report() in the file brainstorm3/toolbox/process/bst_report.m. This is made possible with the use of the short script "macro_methodcall" you will see at the top of many files. Many of the Brainstorm .m files are actually libraries of functions, rather than simple "scripts" or "functions". == Line by line: Body == {{{ % Process: Select data files in: Subject01/*/Avg: deviant sFiles = bst_process('CallProcess', 'process_select_files_data', sFiles, [], ... 'subjectname', SubjectNames{1}, ... 'condition', '', ... 'tag', 'Avg: deviant', ... 'includebad', 0, ... 'includeintra', 0, ... 'includecommon', 0); % Process: Low-pass:30Hz sFiles = bst_process('CallProcess', 'process_bandpass', sFiles, [], ... 'highpass', 0, ... 'lowpass', 30, ... 'mirror', 1, ... 'sensortypes', 'MEG', ... 'overwrite', 0); % Process: Snapshot: Recordings time series sFiles = bst_process('CallProcess', 'process_snapshot', sFiles, [], ... 'target', 5, ... % Recordings time series 'modality', 1, ... % MEG (All) 'orient', 4, ... % bottom 'time', 0.11, ... 'contact_time', [0, 0.1], ... 'contact_nimage', 12, ... 'threshold', 20, ... 'Comment', 'Run'); }}} You will find one block per process you selected in the pipeline editor. They all have the same syntax: <<BR>> output_files = '''bst_process'''('CallProcess', process_name, input_files_A, input_files_B, options_list); * '''process_name''': String indicating the function corresponding to the process to execute. To know from the pipeline editor what is the path to the process function: hover your mouse over the selected process, as illustrated in [[http://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor#Saving_a_pipeline|this tutorial]]. * '''input_files_A''': List of input files in Process1, or FilesA in Process2. It can be a cell array of files names (full path, or relative path from the protocol folder), or an array of structures describing the files in the database (returned by a previous call to bst_process). * '''input_files_B''': Empty for Process1, or FilesB in Process2. Cell array of strings or array of struct. * '''options_list''': Pairs of (option_name, option_values), one for each option of the process. * '''output_files''': Array of structures describing the files in output of the process. If the process created new files, this variable contains the new files. If the process didn't create new files or was modifying exiting files, most of the time this variable would contain the same files as the input list. == Line by line: Footer == {{{ % Save and display report ReportFile = bst_report('Save', sFiles); }}} Closes the current report and saves it in the user Brainstorm report folder ($HOME/.brainstorm/reports). These reports are in .mat format and contain all the information necessary to re-run the execution exactly in the same way, but they not easy to read. The parameter "sFiles" is optional, it indicates what are the files that are considered as the final results of the script. You can remove it without breaking your script: ReportFile = bst_report('Save'); {{{ bst_report('Open', ReportFile); }}} Opens the report viewer to display what happened during the execution. This is equivalent to using the menu [[http://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor#Report_viewer|File > Report viewer]]. You can comment this line (ie. add a "%" at the beginning of the line) if you don't want to show the report at the end of the execution. {{{ % bst_report('Export', ReportFile, ExportDir); }}} This function exports the report in readable format, as an HTML that includes all the screen captures embedded in it. It is disabled by default. If you want to use this feature: remove the "%" at the beginning of the line, and define the variable ExportDir. ExportDir must be a string that defines where to save the HTML report. It can be wither the absolute path to a HTML file (eg. 'C:\Users\myuser\Documents\report_example.html') or just a folder (eg. 'C:\Users\myuser\Documents'). If you enter only a path to a folder, a default file name including the protocol name and a date tag is generated (report_ProtocolName_YYMMDD_HHMMSS.html). == Simplify the calls == This script you generated is like any Matlab script: you can edit it, rename the variables, add tests and loops, etc. The first important thing to understand is how to edit the options or change the inputs/outputs of a process. The script generator uses only one variable for all the file lists ('''sFiles''') and the output process is always the input of the following process. This is usually too restrictive to write a full analysis script: we commonly need to have multiple lists of files or to run two different operations on the same file. Let's consider the first process call, which selects the averages for the Deviant condition in both runs. {{{ sFiles = bst_process('CallProcess', 'process_select_files_data', sFiles, [], ... 'subjectname', SubjectNames{1}, ... 'condition', '', ... 'tag', 'Avg: deviant', ... 'includebad', 0, ... 'includeintra', 0, ... 'includecommon', 0); }}} There is no need to set the parameter sFiles, because there is no input, you can replace it with an empty matrix []. You can therefore remove the line "sFiles = [];". We can also rename the output variable "sAvgData", to be a bit more specific. {{{ sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ... }}} You can omit all the options that are not defined, not used, or kept to their default values: {{{ sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ... 'subjectname', SubjectNames{1}, ... 'tag', 'Avg: deviant'); }}} Edit low-pass filter call: Change the input to sAvgData and the output to sAvgDataLow, this way you will be able to keep track of the two files if you need to use them independently later in the script. {{{ sAvgDataLow = bst_process('CallProcess', 'process_bandpass', sAvgData, [], ... 'highpass', 0, ... 'lowpass', 30, ... 'sensortypes', 'MEG'); }}} Edit the call to the snapshot process: Change the input to sAvgDataLow, and remove the output parameter (we are not expecting output from it). {{{ bst_process('CallProcess', 'process_snapshot', sAvgDataLow, [], ... 'target', 5, ... % Recordings time series 'modality', 1); % MEG (All) }}} Replace the last lines with the following, to export the report instead of opening in the report viewer (edit the file path to point at your own user folder instead). {{{ ReportFile = bst_report('Save'); bst_report('Export', ReportFile, 'C:\Users\franc\Documents\report_test.html'); }}} == Evaluate in Matlab == Select the code for the first process in the Matlab editor, right-click > Evaluate selection (or press F9). {{attachment:edit1.gif}} If you haven't executed your script yet, you will get the following error in the Matlab command window: {{{ Undefined variable "SubjectNames" or class "SubjectNames". }}} The variable SubjectNames is not defined yet: Execute the first two lines "SubjectNames = {'Subject01'}", then try again. You should now have a new variable in your Matlab workspace, which points at the two average files. Type "sAvgData(1)" in your command window to display the first element: {{{ >> sAvgData(1) ans = iStudy: 6 iItem: 1 FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat' FileType: 'data' Comment: 'Avg: deviant (39 files)' Condition: 'S01_AEF_20131218_01_600Hz_notch' SubjectFile: 'Subject01/brainstormsubject.mat' SubjectName: 'Subject01' DataFile: '' ChannelFile: 'Subject01/S01_AEF_20131218_01_600Hz_notch/channel_ctf_acc1.mat' ChannelTypes: {'ADC A' 'ADC V' 'DAC' 'ECG' 'EOG' 'FitErr' 'HLU' 'MEG' 'MEG REF' ...} }}} The field "sAvgData(1).FileName" contains the relative path to the to the Deviant average in the first run. This structure sAvgData contains also a lot of information that can be helpful in your script: * '''iStudy / iItem''': Reference of the file in the database (described later in this tutorial). * '''FileType''': 'raw' (continuous files), 'data' (recordings), 'results' (sources), 'timefreq' (time-frequency, spectrum and connectivity), or 'matrix' (any time series extracted from other files). * '''Comment''': Comment field of the file (what is displayed in the database explorer) * '''Condition''': Name of the condition/folder in which the file is located * '''SubjectFile''': Relative path to the subject file (brainstormsubject.mat) * '''SubjectName''': Name of the subject * '''DataFile''': For types 'results' or 'timefreq', path of the parent file in the database explorer * '''ChannelFile''': Relative path to the channel file * '''ChannelTypes''': Cell array of channel types available for the input file == Naming conventions == To help you navigate in the Brainstorm code, here are some naming conventions: * '''Structures''': Name starting with a "s" followed by a capital letter (eg. sFiles, sStudy, sSubject). * '''Indices''': Either loop variables or array indices, name starting with a "i" (eg. iSubject, iStudy, iTime). * '''Counts''': Number of elements in a group, name starting with a "n" (eg. nAvg, nTrials, nSubjects). * '''File names''': Scripts and functions, only lower case, separation with "_" (eg. process_fft, bst_get). * '''Sub-functions''': Inside a .m file, name starting with a capital, CamelCase (eg. CallProcess, Start). * '''Graphic handles''': Matlab graphic objects, name starting with a "h" (eg. hFig, hAxes, hLine, hText). == Running the script == The simplified script looks like this: {{{ % Input files SubjectNames = {'Subject01'}; % Start a new report bst_report('Start'); % Process: Select data files in: Subject01/*/Avg: deviant sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ... 'subjectname', SubjectNames{1}, ... 'tag', 'Avg: deviant'); % Process: Low-pass:30Hz sAvgDataLow = bst_process('CallProcess', 'process_bandpass', sAvgData, [], ... 'highpass', 0, ... 'lowpass', 30, ... 'sensortypes', 'MEG'); % Process: Snapshot: Recordings time series bst_process('CallProcess', 'process_snapshot', sAvgDataLow, [], ... 'target', 5, ... % Recordings time series 'modality', 1); % MEG (All) % Save and display report ReportFile = bst_report('Save'); bst_report('Export', ReportFile, 'C:\Users\franc\Documents\report_test.html'); }}} You have three ways to execute it: * Select all the lines (Ctrl+A) and evaluate it in Matlab (F9). * In the Editor toolbar of the Matlab environment, click on the button [Run]. * Save the file, go to this folder with Matlab (or add it to your path) and type the name of the script in the command window (without the ".m" at the end). At the end of the execution, nothing happens, because we indicated we wanted to just export the report instead of opening it. To check out the report of execution: use the menu '''File > Report viewer''' from the Brainstorm window, or open the '''report_test.html''' that was saved somewhere on your computer. On this page, you can get review everything that happened in the script: when it was executed, how long it took, what are the processes that were executed, some produced messages (two files were selected with the first process), and at the end you have the screen captures taken by process_snapshot. . {{attachment:report1.gif||height="455",width="593"}} == Running the script again == If you execute the script again, it will not behave as expected anymore. The selection process we used assumes that there is only one file per folder with a comment that includes "Avg: deviant". This is not the case anymore after the execution, because the low-pass filtered files also contain the same tags. The execution of the first process of the script now '''returns 4 files'''. {{{ >> sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ... 'subjectname', SubjectNames{1}, ... 'tag', 'Avg: deviant') sAvgData = 1x4 struct array with fields: iStudy iItem ... }}} In order to exclude the low-pass filtered files from this selection, you can add another process that will '''refine the selection'''. Use the script generator again to create a template call for another process, then copy-paste it in your script. * In Process1: Select any recordings file (we will not run anything, just generate a script). * Select process '''File > Select files: By tag''': Search="low", Search the comment, '''Ignore '''the files. <<BR>><<BR>> {{attachment:select_tag.gif||height="252",width="261"}} * Select the menu '''Generate .m script''' (make sure you do not overwrite the script you are currently working on), then close the pipeline editor. * Copy-paste and edit the call to process_select_tag to your main script. Now the file selection part of your script should look like this, and should return only two files: {{{ % Process: Select data files in: Subject01/*/Avg: Deviant sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ... 'subjectname', SubjectNames{1}, ... 'tag', 'Avg: deviant'); % Process: Ignore file comments with tag: low sAvgData = bst_process('CallProcess', 'process_select_tag', sAvgData, [], ... 'tag', 'low', ... 'search', 2, ... % Search the file comments 'select', 2); % Ignore the files with the tag }}} With this last modification, your script is more robust. It can be executed multiple times without completely changing its behavior. When you are fetching files from the database using comment tags or file names, always pay attention to this aspect: the database grows and the further you go, the more specific your requests have to be. A good practice can be to tag explicitely the output files your script generates if you need to fetch them later. You can use the process '''File > Add tag''' and '''File > Set comment'''. . {{attachment:add_tag.gif||height="203",width="456"}} == Starting Brainstorm == Brainstorm must be running in the background for these scripts to run properly. The interface doesn't have to be visible on the screen, but the database engine must be running for processing requests. At the beginning of your script, you can explicitely start or restart Brainstorm. '''brainstorm''': Start Brainstorm with the regular GUI. '''brainstorm nogui''': Start in silent mode, without the GUI, without progress bar, without user interactions. If you want to start Brainstorm only if Brainstorm is not already running, you can use the following code: {{{ if ~brainstorm('status') brainstorm nogui end }}} To select a specific protocol at the beginning of your script: {{{ ProtocolName = 'TutorialIntroduction'; % Get the protocol index iProtocol = bst_get('Protocol', ProtocolName); if isempty(iProtocol) error(['Unknown protocol: ' ProtocolName]); end % Select the current procotol gui_brainstorm('SetCurrentProtocol', iProtocol); }}} To delete the protocol and start over: {{{ % Delete existing protocol gui_brainstorm('DeleteProtocol', ProtocolName); % Create new protocol gui_brainstorm('CreateProtocol', ProtocolName, 0, 0); }}} == Database requests == The functions '''bst_get''' and '''bst_set''' allow you to query the database, access the configuration of the software and some display parameters. The complete reference documentation of these functions is included directly in their code (brainstorm3/toolbox/core/bst_get.m and bst_set.m). Let's start with a few simple examples: {{{ >> ProtocolInfo = bst_get('ProtocolInfo') % Returns the configuration of the current protocol ProtocolInfo = Comment: 'TutorialIntroduction' STUDIES: 'C:\Work\Protocols\TutorialIntroduction\data' SUBJECTS: 'C:\Work\Protocols\TutorialIntroduction\anat' iStudy: 6 UseDefaultAnat: 0 UseDefaultChannel: 0 >> isGUI = bst_get('isGUI') % Is the Brainstorm interface currently displayed (0=no, 1=yes) >> bst_set('FlipYAxis', 1) % The new figures will have the Y axis flipped >> bst_set('TSDisplayMode', 'butterfly') % The new figures will use a "butterfly" view >> bst_set('FieldTripDir', FieldTripDir) % Set the path where the FieldTrip toolbox is installed }}} To reference the files in the database, each protocol is subdivided in '''Subjects''' (the "anat" folder and containing the MRI, surfaces and atlases) and '''Studies''' (the "data" folder, including the recordings, channel files and all the analyses). Each Study corresponds to a sub-folder (eg. protocol/data/subject01/run01/), and is attached to only one subject. Subjects and Studies are referenced in the protocol with a unique index most of the time kept in variables named '''iSubject''' and '''iStudy'''. The files available in them are also referenced with indices, with variables such as iAnatomy, iSurface, iData, iHeadModel, iResults, iTimefreq. You can see the indices in the database explorer by hovering your mouse over the nodes of the tree: {{attachment:db_indices.gif||height="64",width="715"}} Example: Getting the '''study structure''' from the variable sAvgData, defined in the script: {{{ >> sAvgData(1) ans = iStudy: 6 iItem: 1 ... >> sStudy = bst_get('Study', sAvgData(1).iStudy) % Get a study structure with its index sStudy = Name: 'S01_AEF_20131218_01_600Hz_notch' FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/brainstormstudy.mat' DateOfStudy: '13-May-2016' BrainStormSubject: 'Subject01/brainstormsubject.mat' % Corresponding subject filename Condition: {'S01_AEF_20131218_01_600Hz_notch'} % Name of the condition (=folder) Channel: [1x1 struct] % Channel file for the folder iChannel: [] % Not used anymore Data: [1x242 struct] % List of "data" files in the folder HeadModel: [1x1 struct] % List of head models in the folder iHeadModel: 1 % Default head model (file in green) Result: [1x244 struct] % List of source files and source links Stat: [1x0 struct] % List of statistical results Image: [0x0 struct] % List of images NoiseCov: [1x2 struct] % Noise(1) and data(2) covariance files Dipoles: [0x0 struct] % List of dipole files in the folder Timefreq: [1x247 struct] % List of time-frequency files Matrix: [0x0 struct] % List of "matrix" files in the folder % If 1, the trial is marked as bad in the explorer }}} Example: Getting the '''data structure'''. {{{ >> sData = sStudy.Data(sAvgData(1).iItem) % Get the structure representing the file from sStudy sData = FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat' Comment: 'Avg: deviant (39 files)' % File comment, displayed in the database explorer DataType: 'recordings' % Type of data in the file (mostly "recordings") BadTrial: 0 }}} Example: Getting the '''subject structure'''. {{{ >> sSubject = bst_get('Subject', sStudy.BrainStormSubject) % Get subject structure from filename sSubject = Name: 'Subject01' % Subject name, same as folder name Comments: '' % Not used much FileName: 'Subject01/brainstormsubject.mat' DateOfAcquisition: '' % Not used anymore Anatomy: [1x1 struct] % List of MRI volumes in the subject folder Surface: [1x9 struct] % List of surfaces in the subject folder iAnatomy: 1 % Default MRI volume (in green, index of array Anatomy) iScalp: 9 % Default head surface (index of array Surface) iCortex: 4 % Default cortex surface (index of array Surface) iInnerSkull: [] % Default inner skull surface (index of array Surface) iOuterSkull: [] % Default outer skull surface (index of array Surface) iOther: [] % Not used anymore UseDefaultAnat: 0 % If 1: Subject uses the default anatomy UseDefaultChannel: 0 % 0=one channel/folder, 1=one/subject, 2=one global }}} Example: Getting the '''study structure''' and '''data index''' from a file name. {{{ >> DataFile = sAvgData(1).FileName DataFile = Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat >> [sStudy, iStudy, iData] = bst_get('DataFile', DataFile) sStudy = Name: 'S01_AEF_20131218_01_600Hz_notch' FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/brainstormstudy.mat' DateOfStudy: '13-May-2016' BrainStormSubject: 'Subject01/brainstormsubject.mat' Condition: {'S01_AEF_20131218_01_600Hz_notch'} Channel: [1x1 struct] iChannel: [] Data: [1x242 struct] HeadModel: [1x1 struct] iHeadModel: 1 Result: [1x244 struct] Stat: [1x0 struct] Image: [0x0 struct] NoiseCov: [1x2 struct] Dipoles: [0x0 struct] Timefreq: [1x247 struct] Matrix: [0x0 struct] iStudy = 6 iData = 1 }}} Many other options are available for searching files in the database with bst_get. We cannot list them all in this page, but you can refer to the code of '''bst_get.m''' for more information. <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:350px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?core=bst_get.m")>><<HTML(</div >)>> <<BR>>To change parameters or database structures: '''bst_set.m'''. <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:350px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?core=bst_set.m")>><<HTML(</div >)>> == File structures == The structures of the different types of files functions were described in the sections "On the hard drive" of the introduction tutorials. Here is a directory of all these sections: * '''Continuous recordings''': File type "raw", [[http://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#On_the_hard_drive|Event markers]]. * '''Imported recordings''': File type "data", [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#On_the_hard_drive|Import epochs]] and [[http://neuroimage.usc.edu/brainstorm/Tutorials/Averaging#On_the_hard_drive|Average response]]. * '''Power spectrum''': File type "timefreq", [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#On_the_hard_drive|Power sprectum and frequency filters]]. * '''Channel files''': File type "channel", [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#On_the_hard_drive|Channel file / MEG-MRI coregistration]]. * '''SSP/ICA projectors''': Saved in the channel files, [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#On_the_hard_drive|Artifact cleaning with SSP]]. * '''Head models''': File type "headmodel", [[http://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel#On_the_hard_drive|Head model]]. * '''Noise covariance''': File types "noisecov" and "ndatacov", [[http://neuroimage.usc.edu/brainstorm/Tutorials/NoiseCovariance#On_the_hard_drive|Noise and data covariance matrices.]] * '''Source files''': File types "results" and "link", [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#On_the_hard_drive|Source estimation]]. * '''Time-frequency''': File type "timefreq", [[http://neuroimage.usc.edu/brainstorm/Tutorials/TimeFrequency#On_the_hard_drive|Time-frequency]]. * '''Statistics''': File types "pdata", "presults", "ptimefreq" and "pmatrix", [[http://neuroimage.usc.edu/brainstorm/Tutorials/Statistics#On_the_hard_drive|Statistics]]. * '''MRI volumes''': File type "subjectimage", [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_MRI|Display the anatomy]]. * '''Surfaces''': File type "tess", [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_Surface|Display the anatomy]]. * '''Scouts and atlases''': Saved in the surface files, [[http://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#On_the_hard_drive|Scouts]]. == Custom processing == In many situations, you will find useful to read the files available in the database, and maybe modify them. The easiest approaches do not require any scripting, we will start by reviewing them quickly. === Process: Run Matlab command === If you want to modify the values saved in a file (eg. the field "F" from a "data" file), the easiest way is probably to use the process1 '''File > Run Matlab command'''. It is also available from Process2 in the category "Other". It loads the files in input and run them through a piece of Matlab code that you can edit freely. It can extend a lot the flexibility of the Brainstorm pipeline manager, providing an easy access to any Matlab function or script. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutUserProcess?action=AttachFile&do=get&target=runMatlab.gif|runMatlab.gif|class="attachment"}} The corresponding script looks like this: {{{ sFiles = bst_process('CallProcess', 'process_matlab_eval', sFiles, [], ... 'matlab', 'Data = Data.^2;', ... 'sensortypes', 'MEG', ... 'overwrite', 0); }}} === Export/Import with the database explorer === Right-click on the Deviant average in the database explorer > '''File > Export to Matlab''' > "DataMat". . {{attachment:export_matlab.gif||height="118",width="355"}} {{{ Data exported as "DataMat" >> DataMat DataMat = ChannelFlag: [340x1 double] ColormapType: [] Comment: 'Avg: deviant (39 files)' DataType: 'recordings' Device: 'CTF' DisplayUnits: [] F: [340x361 double] History: {45x3 cell} Std: [] Time: [1x361 double] nAvg: 39 Events: [1x1 struct] }}} DataMat is the exact content of the corresponding .mat file, as loaded with Matlab's load() function. Edit some of the fields of this structure from the Matlab command window: {{{ >> DataMat.Comment = 'Test square'; >> DataMat.F = DataMat.F .^ 2; }}} Now right-click on the '''folder''' containing the original file > '''File > Import from Matlab''' > DataMat: {{attachment:import_matlab.gif||height="74",width="708"}} If instead, you right-click on the original file and select the menu File > Import from Matlab, it overwrites the selected file instead of creating a new one with the selected structure. == Reference: File manipulation == Useful functions for manipulating file names and paths (read the code of the functions for help): * '''file_fullpath''': Converts a relative file path from the Brainstorm database to an absolute path. * '''file_short''': Converts an absolute file path to a short path, relative to the current protocol folder. * '''file_gettype''': Returns the type of a file. Reading files from a script (all the functions take relative paths in input): * '''in_bst_data'''(DataFile): Read an imported epoch. * '''in_bst_timefreq'''(TimefreqFile): Read a power spectrum, time-frequency or connectivity file. * '''in_bst_headmodel'''(HeadmodelFile, ApplyOrient): Read a head model file and apply orientations. * '''in_bst_results'''(ResultsFile, LoadFull): Load a source file and optionally reconstruct the full source time series on the fly (ImagingKernel * recordings). * '''in_bst_matrix'''(MatrixFile): Read a file with the type "matrix". * '''in_bst'''(FileName, TimeWindow): Read any Brainstorm data file with the possibility to load only a specific part of the file. "TimeWindow" is a range of time values in seconds: [tStart, tStop]. * '''bst_process'''(''''LoadInputFile'''', FileName, Target, TimeWindow): The most high-level function for reading data files. "Target" is a string with the list of signal names or types to load. * '''bst_memory('GetConnectMatrix'''', TimefreqMat): Rebuild a full connectivity matrix. * '''in_fopen'''(DataFile, FileFormat, ImportOptions): Low-level function for opening continuous files. * '''in_fread'''(sFile, ChannelMat, iEpoch, SamplesBounds, iChannels, ImportOptions): Low-level function for reading blocks from continuous files. Require a call to in_fopen first to get the sFile structure. Saving files: * '''bst_save'''(FileName, FileMat, Version, isAppend): Save a file but does not register it in the database. * FileName: Must be an absolute path, use in combination with file_fullpath to use relative paths. * FileMat: Must be a valid Brainstorm structure, corresponding to the file type. * Version: Defines which version of the Matlab .mat format is used to store the data: <<BR>>- 'v6': Fastest option, bigger files, no compression, no files >2Gb<<BR>>- 'v7': Slower option, compressed, no files >2Gb<<BR>>- 'v7.3': Much slower than the others, compressed, but only way to save files > 2Gb. * isAppend: If set to 1, updates only the fields defined in FileMat, keep the others untouched. * '''file_unique''': Produces a unique file name by adding a number tag to it. * '''bst_process('GetNewFilename'''', OutputFolder, BaseFilename): Generate a new unique file name based on the beginning of the file name (eg. BaseFilename='data_average_test_'). Registering new files in the database: * '''db_add'''(iStudy/iSubject, FileMat): Add a new file in an anatomy folder or a functional data folder. This function saves and then registers it in the database. FileMat must be a structure, not a filename. You should not save the file manually before calling this function. * '''db_add_data'''(iStudy, FileName, FileMat): Register in the database a structure FileMat that has already been saved in file FileName. You should call bst_save manually before calling this function. * '''db_add_data'''(iStudy, FileName, FileMat, iItem): Overwrites the existing file #iItem. Reload folders (if you saved or deleted files without registering correctly the modification in the database): * '''db_reload_studies'''(iStudies): Reload only the select data folders (aka "studies"). * '''db_reload_conditions'''(iSubjects): Reload all the data folders for a subject. * '''db_reload_subjects'''(iSubjects): Reload the anatomy of the selected subjects. * '''db_reload_database'''('current'): Reload the entire protocol (anatomy and functional data). Other useful database functions: * '''db_add_condition''': Create a new folder in a subject. * '''db_add_subject''': Create a new subject. * '''db_delete_studies''': Delete a list of folders. * '''db_delete_subjects''': Delete a list of subjects. * '''db_group_conditions''': Merge two analysis folders from the same subject. * '''db_rename_condition''': Rename a folder. * '''db_rename_subject''': Rename a subject * '''db_set_channel''': Set the channel file for a folder. * '''db_set_noisecov''': Set the noise/data covariance for a folder, or copy to other folders/subjects. * '''db_set_template''': Copy an anatomy template to a subject or use it as the default anatomy. Export a file from the database to other file formats (read the comments in the functions for help): * export_channel * export_data * export_events * export_result * export_timefreq * export_matrix * export_mri * export_surfaces * export_protocol: Export a subject or an entire protocol as a .zip file. Convert Brainstorm structures to FieldTrip structures: * out_fieldtrip_channel * out_fieldtrip_data * out_fieldtrip_timefreq * out_fieldtrip_headmodel * out_fieldtrip_results * out_fieldtrip_matrix == Reference: Display functions == Create new visualization figures: * '''view_channels''': Display sensors in a 3D figure. * '''view_helmet''': Display the inner surface of the MEG helmet in a 3D figure. * '''view_timeseries''': Display a data file as time series. * '''view_timeseries_matrix''': Display a custom matrix as time series. * '''view_topography''': Display a data file as a spatial topography. * '''view_erpimage''': Display multiple data files as an image, signal by signal. * '''view_timefreq''': Open a time-frequency file (various display modes available). * '''view_spectrum''': Display power spectrum (PSD or time-frequency files). * '''view_connect''': Open a connectivity matrix (various display modes available). * '''view_matrix''': Open a "matrix" file (various display modes available). * '''view_contactsheet''': Create a contact sheet in time or across a volume from an existing figure. * '''view_noisecov''': Display a noise or data covariance file. * '''view_dipoles''': Open a dipoles file (various display modes available). * '''view_pac''': Open PAC results (various display modes available). * '''view_mri''': View a MRI file in the MRI viewer (with or without a functional overlay from a source file). * '''view_mri_3d''': View a MRI file in a 3D figure (with or without a functional overlay from a source file). * '''view_surface''': View a surface. * '''view_surface_data''': View a surface file with a source file as its texture. * '''view_surface_matrix''': View a custorm surface (user defines vertices, faces, color, transparency). * '''view_image_reg''': Display a 3D or 4D matrix as an image with time and/or frequency dimensions. * '''view_struct''': Display a structure as with the popup menu "File > View file contents". * '''script_view_sources''': Shortcut script to display source files. * bst_memory(''''UnloadAll'''', 'Forced'): Close all the existing figures. Configure time-series figures: * panel_time(''''SetCurrentTime'''', t): Set current time. * panel_record(''''SetTimeLength'''', duration): Set the length of the current display page, in seconds. * panel_record(''''SetStartTime'''', t): Set the start of the current page, in seconds. * panel_record(''''SetDisplayMode'''', hFig, DisplayMode): 'column' or 'butterfly' * panel_filter(''''SetFilters'''', isLowPass, LowPass, isHighPass, HighPass, isSinRem, SinRem, isMirror) * panel_montage(''''SetCurrentMontage'''', hFig, MontageName): Change the montage of channels. * bst_figures(''''SetSelectedRows'''', SelectedChannels): Set selected channels (cell array of strings). * figure_timeseries(''''SetTimeSelectionManual'''', hFig, TimeWindow): Select a time segment. Configure 3D figures: * figure_3d(''''SetStandardView'''', hFig, 'left'): Change camera view (top,bottom,left,right,front,back). * figure_3d(''''ViewSensors'''', hFig, isMarkers, isLabels): Enable the view of sensor markers and labels. * panel_surface(''''SetShowSulci'''', hFig, iTess, 1): Enable/Display the display of sulci. * panel_surface(''''SetSurfaceColor'''', hFig, iTess, [1 0 0]): Set surface color. * panel_surface(''''SetSurfaceSmooth'''', hFig, iTess, Smooth, 0): Set amount of smoothing (0-1). * panel_surface(''''SetSurfaceTransparency'''', hFig, iTess, Transp): Set transparency (0-1). * panel_surface(''''SetDataThreshold'''', hFig, iTess, Thresh): Set amplitude threshold. * panel_surface(''''SetSizeThreshold'''', hFig, iTess, MinSize): Set size threshold (min size slider). * figure_mri(''''SetLocation'''', CsName, hFig, [], xyz): CsName=voxel/mri/scs/mni, xyz=[x,y,z] Configure time-frequency figures: * panel_freq(''''SetCurrentFreq'''', iFreq, 1): Set the the current frequency index (from the Freqs vector) * panel_freq(''''SetCurrentFreq'''', Freq, 0): Set the the current frequency (in Hz). * sOptions = panel_display(''''GetDisplayOptions''''): Get display options selected in the Display tab. * panel_display(''''SetDisplayOptions'''', sOptions): Change the options selected in the Display tab. * sOptions.'''HideEdgeEffects''': Controls the checkbox "Hide edge effects" (0 or 1). * sOptions.'''HighResolution''': Controls the checkbox "Smooth display" (0 or 1). * sOptions.'''RowName''': Controls the signal that is currently displayed (for 'SingleSensor' display mode) * sOptions.'''Function''': Controls the display function (magnitude, power, log, phase). Configure colormaps: * bst_colormaps(''''SetColormapName'''', ColormapType, ColormapName): 'jet', 'parula', 'cmap_rbw', ... * bst_colormaps(''''SetMaxMode'''', ColormapType, MaxMode): Colorbar range ('global', 'local', 'custom'). * bst_colormaps(''''SetMaxCustom'''', ColormapType, [], Min, Max): Set a custom colorbar range. * bst_colormaps(''''SetColormapAbsolute'''', ColormapType, isAbsolute): Show positive or relative values * bst_colormaps(''''SetDisplayColorbar'''', ColormapType, isDisplay): Show colorbar in the figures. * bst_colormaps(''''RestoreDefaults'''', ColormapType): Restore the default configuration for a colormap. * ColormapType: anatomy, meg, eeg, sources, stat1, stat2, time, timefreq, connect1, connectn, image Configure statistical thresholding: * StatThreshOptions = bst_get(''''StatThreshOptions''''): Get display options selected in the Stat tab. * bst_set(''''StatThreshOptions'''', StatThreshOptions): Change the options selected in the Stat tab. * StatThreshOptions.'''pThreshold''': Current p-Value threshold. * StatThreshOptions.'''Correction: '''Correction for multiple comparisons ('none', 'fdr', 'bonferroni') * StatThreshOptions.'''Control''': List of dimensions to correct for multiple comparisons (default = [1 2 3]) Export the contents of a figure to a file: * '''out_figure_image''': Screen capture of any Brainstorm figure. * '''out_figure_movie''': Save a movie from one or multitple Brainstorm figures. * '''out_figure_timefreq''': Extract some of the data displayed in a time-frequency figure. * '''out_figure_timeseries''': Extract some of the data displayed in a time series figure. == Example: Creating a new file == This section illustrates how to add new files to the database. We will create a sinusoidal signal and save it in a "matrix" file, in a new folder of subject Test. {{{ % Create two sinsuoidal signals (20Hz,30Hz), over 1 second with a sampling frequency of 1000Hz t = 0:0.001:1; F = [sin(20*2*pi*t); 0.5*sin(30*2*pi*t)]; % Initialize an empty "matrix" structure sMat = db_template('matrixmat'); % Fill the required fields of the structure sMat.Value = F; sMat.Comment = 'Test sinusoids'; sMat.Description = {'Signal #1: 20Hz'; 'Signal #2: 30Hz'}; sMat.Time = t; % Create a new folder "Script" in subject "Test" iStudy = db_add_condition('Test', 'Script'); % Get the corresponding study structure sStudy = bst_get('Study', iStudy); }}} There are many options to add a new file to the database, with various levels of requirements. You can call the db_add function (reloads the destination folder, therefore slow if you save many files), save the file in the corresponding folder and reload the protocol, or register the file in the database manually (more complicated but faster). '''Option #1: db_add''' {{{ OutputFile = db_add(iStudy, sMat); }}} '''Option #2: bst_save / '''db_reload_studies {{{ % Another to generate a unique filename (without a timestamp) MatrixFile = file_unique(bst_fullfile(OutputFolder, 'matrix_test.mat')); % Save file bst_save(MatrixFile, sMat, 'v6'); % Reload the folder in which the new file was saved db_reload_studies(iStudy); }}} '''Option #3: bst_save / '''db_add_data {{{ % Get the full path to the new folder (same folder as the brainstormstudy.mat file for this study) OutputFolder = bst_fileparts(file_fullpath(sStudy.FileName)); % Get a new unique filename (including a timestamp) MatrixFile = bst_process('GetNewFilename', OutputFolder, 'matrix_test'); % Save file bst_save(MatrixFile, sMat, 'v6'); % Reference saved file in the database db_add_data(iStudy, MatrixFile, sMat); % Update tree panel_protocols('UpdateNode', 'Study', iStudy); }}} {{attachment:newfile.gif||height="166",width="521"}} == Example: Editing events == A step that commonly requires manual changes is the definition of the event markers. Many time we have to combine external triggers or behavioral information with the existing events. This example illustrates how to load the events, modify them and save them back. For the continuous recordings, the events are saved in the .mat file corresponding to the "Link to raw file" available in the database explorer. These structures contain only meta-data and information created with Brainstorm, the EEG/MEG recordings are available in a separate binary file. First, we need to load this link. {{{ % Right-click on a "Link to raw file" in the database explorer > File > Copy file path to clipboard RawFile = '/.../@rawS01_AEF_20131218_01_600Hz_notch/data_0raw_S01_AEF_20131218_01_600Hz_notch.mat' % Load the "sFile" structure, contained in the .F structure of the link .mat file sRaw = in_bst_data(RawFile, 'F'); >> sRaw.F.events ans = 1x7 struct array with fields: label color epochs samples times reactTimes select }}} For example, let's say we want to add 30ms to all the events in the category "button", to compensate for some hardware delay, and create a new event category with the modified timing. We need first to identify what is the '''index of the category "button"''', in this array of 7 events structures. {{{ % Find the index of the event category "button" iEvtButton = find(strcmpi({sRaw.F.events.label}, 'button')); >> iEvtButton iEvtButton = 3 }}} In the code above, note this special Matlab syntax that allows the '''concatenation''' of the values of one field across multiple structures, in an array of structures: {{{ >> {sRaw.F.events.label} ans = 'standard' 'deviant' 'button' 'cardiac' 'blink' 'bad_1-7Hz' 'bad_40-240Hz' }}} If you wanted to search instead all the events containing a specific tag, for example "bad", you can use the '''cellfun''' function (function that applies the same function sequentially to all the elements in a cell array and concatenates the results) in combination with the '''strfind''' function (search for a substring). The final call to the '''find''' function returns at which indices the list of tags found in the event label is not empty. {{{ >> iEvtBad = find(~cellfun(@(c)isempty(strfind(c,'bad')), {sRaw.F.events.label})) iEvtBad = 6 7 }}} The code below copies the existing event category "button", renames it and add a 30ms offset. Note that '''times''' and '''samples''' field must always match. If you modify one, you must also modify the others (times = samples/sfreq). If you add or remove events, you must adjust the size of the '''epochs''' field (always 1 for most file formats). {{{ % Copy the event category "button" to a new cagory iEvtNew = length(sRaw.F.events) + 1; sRaw.F.events(iEvtNew) = sRaw.F.events(iEvtButton); % Rename new event to "button_offset" sRaw.F.events(iEvtNew).label = 'button_offset'; % Compute how many samples correspond to 30ms (18 samples at 600Hz = 30ms) offsetSample = round(0.030 .* sRaw.F.prop.sfreq); % Apply this offset to the events in the "button_offset" category sRaw.F.events(iEvtNew).samples = sRaw.F.events(iEvtNew).samples + offsetSample; % Re-compute the corresponding times sRaw.F.events(iEvtNew).times = sRaw.F.events(iEvtNew).samples ./ sRaw.F.prop.sfreq; % Re-generate an epochs field with only ones (optional here, as we didn't change the number of evt) sRaw.F.events(iEvtNew).epochs = ones(1, size(sRaw.F.events(iEvtNew).times, 2)); % Change the event color to yellow (red=1, green=1, blue=0) sRaw.F.events(iEvtNew).color = [1 1 0]; >> sRaw.F.events(iEvtNew) ans = label: 'button_offset' color: [1 1 0] epochs: [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1] samples: [1x40 double] times: [1x40 double] reactTimes: [] select: 1 }}} The last step is to '''save the modifications''' back to the "Link to raw file". Here the call to '''file_fullpath''' is optional because the variable RawFile was already containing the absolute path to the file. {{{ % Update the sRaw structure to the RawFile file (the last parameter appends to the existing struct) bst_save(file_fullpath(RawFile), sRaw, 'v6', 1); }}} Open the recordings to make sure your transformation worked the way you expected. . {{attachment:edit_events.gif||height="146",width="461"}} == Find examples in the code == The easier way to understand how to use a function is to search the code with the "'''Find files'''" interface in Matlab. Go to the brainstorm3 folder, click on "File files" (or Ctrl+Shift+F), enter the name of function your are looking for in "Find files containing text", Include subfolders, Match case. It will return all the lines that include the string you entered across all the files in Brainstorm. Just double-click on a line to jump to the code in the Matlab editor. {{attachment:find_files.gif||height="307",width="638"}} You can use the same interface to '''find what function is called''' when you click on a button or menu in the interface. Search for the label or the tooltip of the interface element in the same way. The example below shows how to track what happens when you click on the headmodel popup menu "Check spheres". {{attachment:find_files2.gif||height="95",width="726"}} If you have trouble understanding how to set some input parameters, you can use the '''debugger''' to explore a real use case. Place a breakpoint at the begging of your function of interest (watch [[https://www.youtube.com/watch?v=PdNY9n8lV1Y|this tutorial]] if you don't know how to do it), for in example view_timeseries.m. Then click on the corresponding menus in the Brainstorm interface (eg. double-click on on a data file). When the execution reaches the line you selected, it stops and give you back the commands. You can explore the values in all the variables, modify them, and execute the code step by step (many options available in the Editor tab of Matlab). {{attachment:debugger.gif||height="268",width="509"}} == Additional quality control [TODO] == You cann add in the reports all the information that will help you control the quality of the analysis, or even figures you want to include in publications or clinical reports. Create figures and snapshot them {{{ % Display raster plot hFigRaster = view_erpimage({sFilesEpochs1.FileName}, 'erpimage', 'MEG'); bst_report('Snapshot', hFigRaster, sFilesEpochs1(1).FileName, 'ERP image'); }}} Add messages: warnings and errors: == Loop over subject and runs [TODO] == Creating loops is not supported yet by the script generator, but relatively easy to do from a script without having to know too much about Matlab programming. 1) Fill the cell array SubjectNames with all your subjects names, with the same dimensions as the list of input raw files (sFiles) . 2) Add a "for" loop that includes all the bst_process() calls (leave the bst_report() calls and input definition outside) 3) Inside the loop, replace SubjectNames with SubjectNames{i} and sFiles with sFiles(i) == How to process an entire study == This section proposes a standard workflow for processing a full group study with Brainstorm. It contains the same steps of analysis as the introduction tutorials, but separating what can be done automatically from what should be done manually. This workflow can be adapted to most ERP studies (stimulus-based). * '''Prototype''': Start by processing one or two subjects completely '''interactively''' (exactly like in the introduction tutorials). Use the few pilot subjects that you have for your study to prototype the analysis pipeline and check manually all the intermediate stages. Take notes of what you're doing along the way, so that you can later write a script that reproduces the same operations. |
| Line 17: | Line 856: |
| * Run FreeSurfer or BrainSuite to get cortical surfaces and atlases for all the subjects. * Use the menu '''File > Batch MRI fiducials''': It prompts for the selection of the fiducials for all the subjects and saves a file '''fiducials.m''' in each segmentation folder. You will not have to redo this even if you have to start over your analysis from the beginning. * Create a script that calls the process "Import anatomy folder" in a loop for all the subjects. * Alternatives: Create and import the subjects one by one and set the fiducials at the import time. Or use the default anatomy for all the subjects (or use [[Tutorials/TutWarping|warped templates]]). * '''Script 1''': Pre-processing: Loop on the subjects and the acquisition runs. * '''Create link to raw files''': Link all the acquisition runs and the noise recordings to the database |
* '''Segmentation''': Run FreeSurfer/BrainSuite to get surfaces and atlases for all the subjects. * '''File > Batch MRI fiducials''': This menu prompts for the selection of the fiducials for all the subjects and saves a file __fiducials.m__ in each segmentation folder. You will not have to redo this even if you have to start over your analysis from the beginning. * '''Script''': Write a loop that calls the process "Import anatomy folder" for all the subjects. * '''Alternatives''': Create and import the subjects one by one and set the fiducials at the import time. Or use the default anatomy for all the subjects (or use [[Tutorials/TutWarping|warped templates]]). * '''Script #1''': Pre-processing: Loop on the subjects and the acquisition runs. * '''Create link to raw files''': Link the subject and noise recordings to the database. |
| Line 25: | Line 864: |
| * '''Evaluation''': Power spectrum density on all the recordings to evaluate their quality. | * '''Evaluation''': Power spectrum density of the recordings to evaluate their quality. |
| Line 27: | Line 866: |
| * '''Evaluation''': Power spectrum density on all the recordings to make sure the filters worked well | * '''Evaluation''': Power spectrum density of the recordings to make sure the filters worked well. |
| Line 29: | Line 868: |
| * '''Detect artifacts''': Detect heartbeats, Detect eye blinks, Remove simultaneous | * '''Detect artifacts''': Detect heartbeats, Detect eye blinks, Remove simultaneous. |
| Line 31: | Line 870: |
| * '''Compute ICA''': If you have some types of artifacts you'd like to remove with ICA. | * '''Compute ICA''': If you have some artifacts you'd like to remove with ICA (no default selection). |
| Line 33: | Line 872: |
| * '''Export the report to HTML''': One report per subject, or one report for all the subjects. * '''Manual inspection''': For each run. * '''Check the reports''': Information messages (number of events, errors and warnings) and screen captures (registration problems, especially obvious noisy channels, incorrect SSP topographies) * Mark the bad channels * '''Fix the SSP''': For the suspicious runs, open the file viewer, adjust the list of blink and cardiac events, remove and recompute the SSP decompositions, manually select the components. * '''Detect other artifacts''': Run the process on all the runs of all the subjects at once * '''Mark bad segments''': Review the detected artifacts, keep only the ones you want to remove, and then mark the event type as BAD. Review quickly the rest of the file and check that there are no other important artifacts. * '''Additional SSP''': If you find one type of artifact that repeats (typically saccades and SQUID jumps), you can create additional SSP projectors, either with the "SSP: Generic" or directly from the topographies (right-click on the topography figure > Create SSP). * '''Script 2''': Epoching, averaging, sources, time-frequency, etc. * Importing * Averaging * Noise covariance: * Head model * Sources * Time-frequency * Statistics * '''Screenshots''': Averages (time series, topographies, sources at one time point). * Manual inspection: * Check the reports: Check the number of imported epochs in each condition, check the averaged * Regions of interest: If not using predefined regions from an atlas. * Script 3: ROI-based analysis, additional statistics. == Script generation == http://neuroimage.usc.edu/brainstorm/Tutorials/PipelineEditor == Script edition == - Add loops, load files, ... Loops: http://neuroimage.usc.edu/forums/showthread.php?2429-Problem-using-tags == File manipulation == * Modify a structure manually: Export to Matlab/Import from Matlab * File manipulation: file_short, file_fullpath, in_bst_*... * Documentation of all file structures: point at the appropriate tutorials * Select files from the database (with bst_get and processes) === Export as script === Use the menu "Generate .m script" to create a Matlab script that would have the exact same result as running this analysis pipeline from the Brainstorm interface. This script is also available in the Brainstorm distribution: '''brainstorm3/toolbox/script/tutorial_raw.m ''' == Report viewer == Click on Run to start the script. As this process is taking screen captures, do not use your computer for something else at the same time: if another window covers the Brainstorm figures, it will not capture the right images. At the end, the report viewer is opened to show the status of all the processes, the information messages, the list of input and output files, and the screen captures. The report is saved in your home folder ($home/.brainstorm/reports). If you close this window, you can get it back with the menu File > Report viewer. |
* '''Export the report''': One report per subject, or one report for all the subjects, saved in HTML. * '''Manual inspection #1''': * '''Check the reports''': Information messages (number of events, errors and warnings) and screen captures (registration problems, obvious noisy channels, incorrect SSP topographies). * '''Mark bad channels''': Open the recordings, select the channels and mark them as bad. Or use the process "Set bad channels" to mark the same bad channels in multiple files. * '''Fix the SSP/ICA''': For the suspicious runs: Open the file, adjust the list of blink and cardiac events, remove and recompute the SSP decompositions, manually select the components. * '''Detect other artifacts''': Run the process on all the runs of all the subjects at once (select all the files in Process1 and run the process, or generate the equivalent script). * '''Mark bad segments''': Review the artifacts detected in 1-7Hz and 40-240Hz, keep only the ones you really want to remove, then mark the event categories as bad. Review quickly the rest of the file and check that there are no other important artifacts. * '''Additional SSP''': If you find one type of artifact that repeats (typically saccades and SQUID jumps), you can create additional SSP projectors, either with the process "SSP: Generic" or directly from a topography figure (right-click on the figure > Snapshot> Use as SSP projector). * '''Script #2''': Subject-level analysis: Epoching, averaging, sources, time-frequency. * '''Importing''': Process "Import MEG/EEG: Events" and "Pre-process > Remove DC offset". * '''Averaging''': Average trials by run, average runs by subject (registration problem in MEG). * '''Noise covariance''': Compute from empty room or resting recordings, copy to other folders. * '''Head model''': Compute for each run, or compute once and copy if the runs are co-registered. * '''Sources''': Compute for each run, average across runs and subjects in source space for MEG. * '''Time-frequency''': Computation with Hilbert transform or Morlet wavelets, then normalize. * '''Screenshots''': Check the quality of all the averages (time series, topographies, sources). * '''Export the report''': One report per subject, or one report for all the subjects, saved in HTML. * '''Manual inspection #2''': * '''Check the reports''': Check the number of epochs imported and averaged in each condition, check the screen capture of the averages (all the primary responses should be clearly visible). * '''Regions of interest''': If not using predefined regions from an atlas, define the scouts on the anatomy of each subject (or on the template and then project them to the subjects). * '''Script #3''': Group analysis, ROI-based analysis, etc. * '''Averaging''': Group averages for the sensor data, the sources and the time-frequency maps. * '''Statistics''': Contrast between conditions or groups of subjects. * '''Regions of interest''': Any operation that involve scouts. == Final script == The following script from the Brainstorm distribution reproduces the introduction tutorials ("Get started"): '''brainstorm3/toolbox/script/tutorial_introduction.m''' - Report: [[http://neuroimage.usc.edu/bst/examples/report_TutorialIntroduction.html|report_TutorialIntroduction.html]] <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:400px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_introduction.m")>><<HTML(</div >)>> <<BR>>For an example of a script illustrating how to create loops, look at the tutorial [[Tutorials/VisualSingle|MEG visual: single subject]]. '''brainstorm3/toolbox/script/tutorial_visual_single.m''' - Report: [[http://neuroimage.usc.edu/bst/examples/report_TutorialVisual_sub001.html|report_TutorialVisual_sub001.html]] <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:400px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_visual_single.m")>><<HTML(</div >)>> == Additional documentation == * Tutorial: [[Tutorials/TutUserProcess|How to write your own process]] |
Tutorial 28: Scripting
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
The previous tutorials explained how to use Brainstorm in an interactive way to process one subject with two acquisition runs. In the context of a typical neuroimaging study, you may have tens or hundreds of subjects to process in the same way, it is unrealistic to do everything manually. Some parts of the analysis can be processed in batches with no direct supervision, others require more attention. This tutorial introduces tools and tricks that will help you assemble an efficient analysis pipeline.
Requirements: You need a license for the Matlab environment in order to use these tools and execute custom scripts. If you are running the compiled version of Brainstorm with the MCR library, the only custom code you can run is through the menu File > Matlab console and the process "Run Matlab command".
Contents
- Starting a new script
- Line by line: Header
- Line by line: Body
- Line by line: Footer
- Simplify the calls
- Evaluate in Matlab
- Naming conventions
- Running the script
- Running the script again
- Starting Brainstorm
- Database requests
- File structures
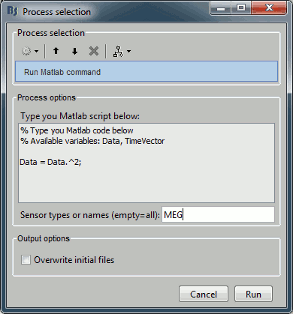
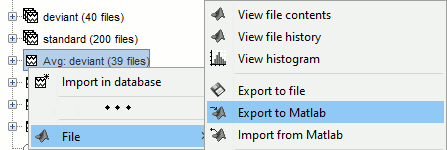
- Custom processing
- Reference: File manipulation
- Reference: Display functions
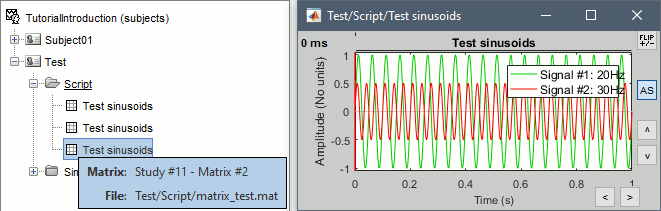
- Example: Creating a new file
- Example: Editing events
- Find examples in the code
- Additional quality control [TODO]
- Loop over subject and runs [TODO]
- How to process an entire study
- Final script
- Additional documentation
Starting a new script
The easiest way to get started with a new Brainstorm script is to use the script generator, already introduced in the tutorial Select files and run processes. Select some files in the Process1 or Process2 tabs, select a list of processes, and use the menu Generate .m script. The example below should work on the the protocol "TutorialIntroduction" created during the introduction tutorials.
- In the Process1 tab, leave the selection box empty and click on [Run]. Instead of selecting the files from the Brainstorm interface, we will select them directly from the database using a script.
Select process File > Select files: Recordings (do not execute immediately)
Subject=Subject01, Condition=[Empty], File comment=Avg: deviant (the space is important).
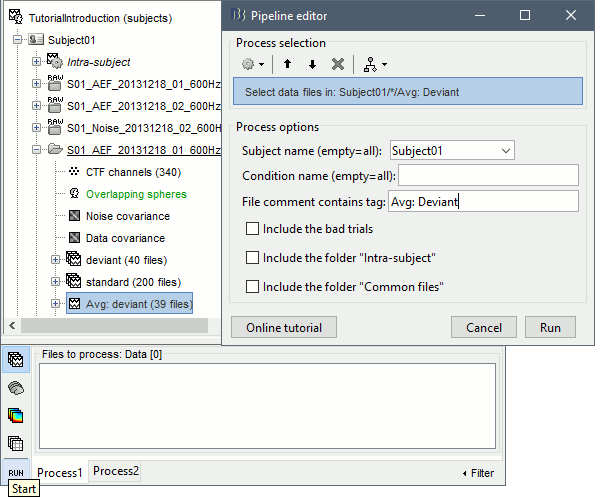
- This process selects all the recordings with a comment including the string "Avg: deviant", from all the folders in Subject01 (except "Intra-subject" and "Common files"). We expect to get two files: the averages of the deviant condition for both runs.
Add process Pre-process > Band-pass filter: Lower cutoff=0Hz, Upper cutoff=30Hz, Mirror.
Add process File > Save snapshot: Recordings time series, Sensors=MEG.
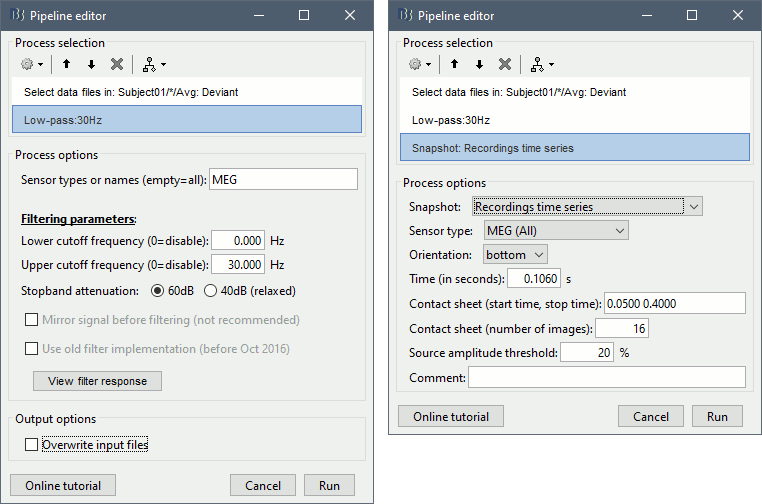
- This will apply a low-pass filter at 30Hz and save a screen capture of the signals in the report.
Do not run the pipeline, select the menu Generate .m script instead. It saves a new .m file and opens it in the Matlab editor. Close the pipeline editor window and look at the script.
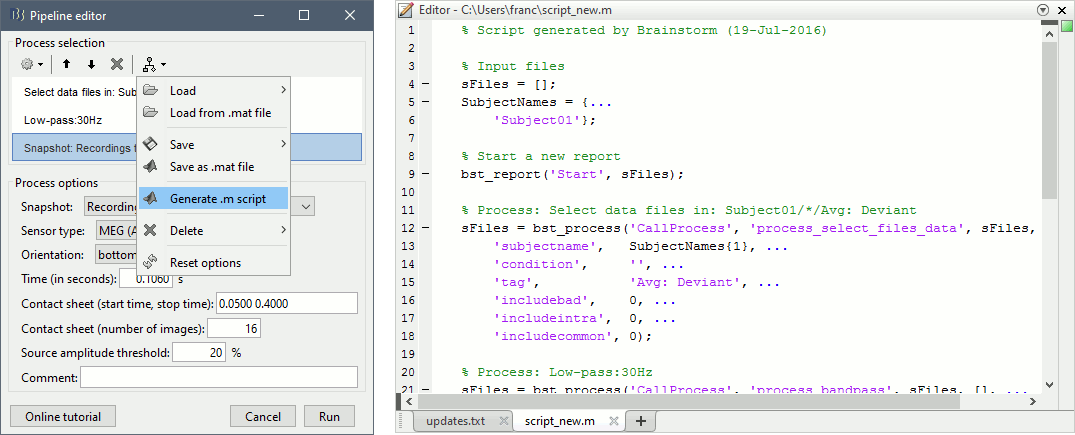
- The script you just generated can be the starting point to your own custom script. The following sections explain line by line how they work and how to edit them.
Line by line: Header
% Script generated by Brainstorm (19-Jul-2016)
All the lines starting with a "%" are comments, they are never executed.
% Input files
sFiles = [];
SubjectNames = {...
'Subject01'};These lines define the script inputs:
sFiles: The list of files in input. Currently empty because we did not select anything in input in the Process1 list. If we had selected files, it would contain a cell array of strings with relative file paths.
SubjectNames: List of subject names that are used in the script. Most of the times, the generated script would contain only one entry, but it written as a cell array so that it is easier to extend it to multiple subjects with a loop (described further in this tutorial).
% Start a new report
bst_report('Start', sFiles);Start a new report of activity: Clears all the previous logs and gets ready to record new messages. The report will collect all the messages that are generated during the execution of the script by the various processes. You can explicitely add screen captures and additional messages to the current report with the function bst_report. This report will remain open until the function bst_report('Start') is called again. To display the current report, use the menu File > Report viewer.
The syntax function_name('SubFunction', arguments) is used a lot in Brainstorm, to call a subfunction available inside a .m file. This line above calls the function Report() in the file brainstorm3/toolbox/process/bst_report.m. This is made possible with the use of the short script "macro_methodcall" you will see at the top of many files. Many of the Brainstorm .m files are actually libraries of functions, rather than simple "scripts" or "functions".
Line by line: Body
% Process: Select data files in: Subject01/*/Avg: deviant
sFiles = bst_process('CallProcess', 'process_select_files_data', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'tag', 'Avg: deviant', ...
'includebad', 0, ...
'includeintra', 0, ...
'includecommon', 0);
% Process: Low-pass:30Hz
sFiles = bst_process('CallProcess', 'process_bandpass', sFiles, [], ...
'highpass', 0, ...
'lowpass', 30, ...
'mirror', 1, ...
'sensortypes', 'MEG', ...
'overwrite', 0);
% Process: Snapshot: Recordings time series
sFiles = bst_process('CallProcess', 'process_snapshot', sFiles, [], ...
'target', 5, ... % Recordings time series
'modality', 1, ... % MEG (All)
'orient', 4, ... % bottom
'time', 0.11, ...
'contact_time', [0, 0.1], ...
'contact_nimage', 12, ...
'threshold', 20, ...
'Comment', 'Run');You will find one block per process you selected in the pipeline editor. They all have the same syntax:
output_files = bst_process('CallProcess', process_name, input_files_A, input_files_B, options_list);
process_name: String indicating the function corresponding to the process to execute. To know from the pipeline editor what is the path to the process function: hover your mouse over the selected process, as illustrated in this tutorial.
input_files_A: List of input files in Process1, or FilesA in Process2. It can be a cell array of files names (full path, or relative path from the protocol folder), or an array of structures describing the files in the database (returned by a previous call to bst_process).
input_files_B: Empty for Process1, or FilesB in Process2. Cell array of strings or array of struct.
options_list: Pairs of (option_name, option_values), one for each option of the process.
output_files: Array of structures describing the files in output of the process. If the process created new files, this variable contains the new files. If the process didn't create new files or was modifying exiting files, most of the time this variable would contain the same files as the input list.
Line by line: Footer
% Save and display report
ReportFile = bst_report('Save', sFiles);Closes the current report and saves it in the user Brainstorm report folder ($HOME/.brainstorm/reports). These reports are in .mat format and contain all the information necessary to re-run the execution exactly in the same way, but they not easy to read.
The parameter "sFiles" is optional, it indicates what are the files that are considered as the final results of the script. You can remove it without breaking your script: ReportFile = bst_report('Save');
bst_report('Open', ReportFile);Opens the report viewer to display what happened during the execution. This is equivalent to using the menu File > Report viewer. You can comment this line (ie. add a "%" at the beginning of the line) if you don't want to show the report at the end of the execution.
% bst_report('Export', ReportFile, ExportDir);This function exports the report in readable format, as an HTML that includes all the screen captures embedded in it. It is disabled by default. If you want to use this feature: remove the "%" at the beginning of the line, and define the variable ExportDir.
ExportDir must be a string that defines where to save the HTML report. It can be wither the absolute path to a HTML file (eg. 'C:\Users\myuser\Documents\report_example.html') or just a folder (eg. 'C:\Users\myuser\Documents'). If you enter only a path to a folder, a default file name including the protocol name and a date tag is generated (report_ProtocolName_YYMMDD_HHMMSS.html).
Simplify the calls
This script you generated is like any Matlab script: you can edit it, rename the variables, add tests and loops, etc. The first important thing to understand is how to edit the options or change the inputs/outputs of a process. The script generator uses only one variable for all the file lists (sFiles) and the output process is always the input of the following process. This is usually too restrictive to write a full analysis script: we commonly need to have multiple lists of files or to run two different operations on the same file.
Let's consider the first process call, which selects the averages for the Deviant condition in both runs.
sFiles = bst_process('CallProcess', 'process_select_files_data', sFiles, [], ...
'subjectname', SubjectNames{1}, ...
'condition', '', ...
'tag', 'Avg: deviant', ...
'includebad', 0, ...
'includeintra', 0, ...
'includecommon', 0);There is no need to set the parameter sFiles, because there is no input, you can replace it with an empty matrix []. You can therefore remove the line "sFiles = [];". We can also rename the output variable "sAvgData", to be a bit more specific.
sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...You can omit all the options that are not defined, not used, or kept to their default values:
sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
'subjectname', SubjectNames{1}, ...
'tag', 'Avg: deviant');Edit low-pass filter call: Change the input to sAvgData and the output to sAvgDataLow, this way you will be able to keep track of the two files if you need to use them independently later in the script.
sAvgDataLow = bst_process('CallProcess', 'process_bandpass', sAvgData, [], ...
'highpass', 0, ...
'lowpass', 30, ...
'sensortypes', 'MEG');Edit the call to the snapshot process: Change the input to sAvgDataLow, and remove the output parameter (we are not expecting output from it).
bst_process('CallProcess', 'process_snapshot', sAvgDataLow, [], ...
'target', 5, ... % Recordings time series
'modality', 1); % MEG (All)Replace the last lines with the following, to export the report instead of opening in the report viewer (edit the file path to point at your own user folder instead).
ReportFile = bst_report('Save');
bst_report('Export', ReportFile, 'C:\Users\franc\Documents\report_test.html');
Evaluate in Matlab
Select the code for the first process in the Matlab editor, right-click > Evaluate selection (or press F9).

If you haven't executed your script yet, you will get the following error in the Matlab command window:
Undefined variable "SubjectNames" or class "SubjectNames".
The variable SubjectNames is not defined yet: Execute the first two lines "SubjectNames = {'Subject01'}", then try again. You should now have a new variable in your Matlab workspace, which points at the two average files. Type "sAvgData(1)" in your command window to display the first element:
>> sAvgData(1)
ans =
iStudy: 6
iItem: 1
FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat'
FileType: 'data'
Comment: 'Avg: deviant (39 files)'
Condition: 'S01_AEF_20131218_01_600Hz_notch'
SubjectFile: 'Subject01/brainstormsubject.mat'
SubjectName: 'Subject01'
DataFile: ''
ChannelFile: 'Subject01/S01_AEF_20131218_01_600Hz_notch/channel_ctf_acc1.mat'
ChannelTypes: {'ADC A' 'ADC V' 'DAC' 'ECG' 'EOG' 'FitErr' 'HLU' 'MEG' 'MEG REF' ...}The field "sAvgData(1).FileName" contains the relative path to the to the Deviant average in the first run. This structure sAvgData contains also a lot of information that can be helpful in your script:
iStudy / iItem: Reference of the file in the database (described later in this tutorial).
FileType: 'raw' (continuous files), 'data' (recordings), 'results' (sources), 'timefreq' (time-frequency, spectrum and connectivity), or 'matrix' (any time series extracted from other files).
Comment: Comment field of the file (what is displayed in the database explorer)
Condition: Name of the condition/folder in which the file is located
SubjectFile: Relative path to the subject file (brainstormsubject.mat)
SubjectName: Name of the subject
DataFile: For types 'results' or 'timefreq', path of the parent file in the database explorer
ChannelFile: Relative path to the channel file
ChannelTypes: Cell array of channel types available for the input file
Naming conventions
To help you navigate in the Brainstorm code, here are some naming conventions:
Structures: Name starting with a "s" followed by a capital letter (eg. sFiles, sStudy, sSubject).
Indices: Either loop variables or array indices, name starting with a "i" (eg. iSubject, iStudy, iTime).
Counts: Number of elements in a group, name starting with a "n" (eg. nAvg, nTrials, nSubjects).
File names: Scripts and functions, only lower case, separation with "_" (eg. process_fft, bst_get).
Sub-functions: Inside a .m file, name starting with a capital, CamelCase (eg. CallProcess, Start).
Graphic handles: Matlab graphic objects, name starting with a "h" (eg. hFig, hAxes, hLine, hText).
Running the script
The simplified script looks like this:
% Input files
SubjectNames = {'Subject01'};
% Start a new report
bst_report('Start');
% Process: Select data files in: Subject01/*/Avg: deviant
sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
'subjectname', SubjectNames{1}, ...
'tag', 'Avg: deviant');
% Process: Low-pass:30Hz
sAvgDataLow = bst_process('CallProcess', 'process_bandpass', sAvgData, [], ...
'highpass', 0, ...
'lowpass', 30, ...
'sensortypes', 'MEG');
% Process: Snapshot: Recordings time series
bst_process('CallProcess', 'process_snapshot', sAvgDataLow, [], ...
'target', 5, ... % Recordings time series
'modality', 1); % MEG (All)
% Save and display report
ReportFile = bst_report('Save');
bst_report('Export', ReportFile, 'C:\Users\franc\Documents\report_test.html');You have three ways to execute it:
- Select all the lines (Ctrl+A) and evaluate it in Matlab (F9).
- In the Editor toolbar of the Matlab environment, click on the button [Run].
- Save the file, go to this folder with Matlab (or add it to your path) and type the name of the script in the command window (without the ".m" at the end).
At the end of the execution, nothing happens, because we indicated we wanted to just export the report instead of opening it. To check out the report of execution: use the menu File > Report viewer from the Brainstorm window, or open the report_test.html that was saved somewhere on your computer.
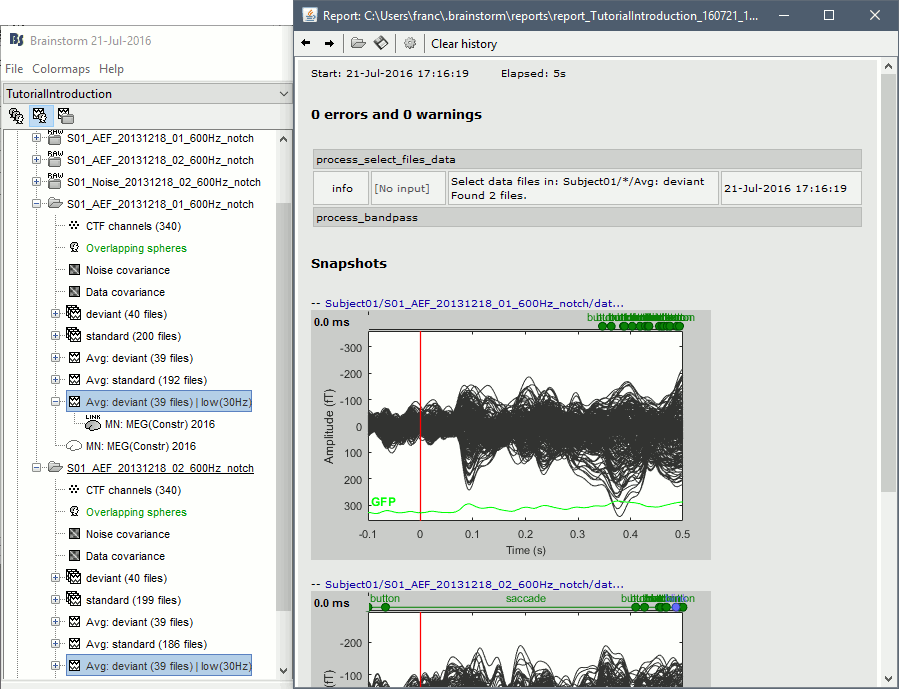
On this page, you can get review everything that happened in the script: when it was executed, how long it took, what are the processes that were executed, some produced messages (two files were selected with the first process), and at the end you have the screen captures taken by process_snapshot.
Running the script again
If you execute the script again, it will not behave as expected anymore. The selection process we used assumes that there is only one file per folder with a comment that includes "Avg: deviant". This is not the case anymore after the execution, because the low-pass filtered files also contain the same tags. The execution of the first process of the script now returns 4 files.
>> sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
'subjectname', SubjectNames{1}, ...
'tag', 'Avg: deviant')
sAvgData =
1x4 struct array with fields:
iStudy
iItem
...In order to exclude the low-pass filtered files from this selection, you can add another process that will refine the selection. Use the script generator again to create a template call for another process, then copy-paste it in your script.
- In Process1: Select any recordings file (we will not run anything, just generate a script).
Select process File > Select files: By tag: Search="low", Search the comment, Ignore the files.
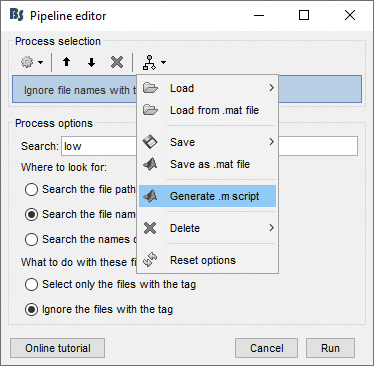
Select the menu Generate .m script (make sure you do not overwrite the script you are currently working on), then close the pipeline editor.
- Copy-paste and edit the call to process_select_tag to your main script.
Now the file selection part of your script should look like this, and should return only two files:
% Process: Select data files in: Subject01/*/Avg: Deviant
sAvgData = bst_process('CallProcess', 'process_select_files_data', [], [], ...
'subjectname', SubjectNames{1}, ...
'tag', 'Avg: deviant');
% Process: Ignore file comments with tag: low
sAvgData = bst_process('CallProcess', 'process_select_tag', sAvgData, [], ...
'tag', 'low', ...
'search', 2, ... % Search the file comments
'select', 2); % Ignore the files with the tagWith this last modification, your script is more robust. It can be executed multiple times without completely changing its behavior. When you are fetching files from the database using comment tags or file names, always pay attention to this aspect: the database grows and the further you go, the more specific your requests have to be.
A good practice can be to tag explicitely the output files your script generates if you need to fetch them later. You can use the process File > Add tag and File > Set comment.
Starting Brainstorm
Brainstorm must be running in the background for these scripts to run properly. The interface doesn't have to be visible on the screen, but the database engine must be running for processing requests. At the beginning of your script, you can explicitely start or restart Brainstorm.
brainstorm: Start Brainstorm with the regular GUI.
brainstorm nogui: Start in silent mode, without the GUI, without progress bar, without user interactions.
If you want to start Brainstorm only if Brainstorm is not already running, you can use the following code:
if ~brainstorm('status')
brainstorm nogui
endTo select a specific protocol at the beginning of your script:
ProtocolName = 'TutorialIntroduction';
% Get the protocol index
iProtocol = bst_get('Protocol', ProtocolName);
if isempty(iProtocol)
error(['Unknown protocol: ' ProtocolName]);
end
% Select the current procotol
gui_brainstorm('SetCurrentProtocol', iProtocol);To delete the protocol and start over:
% Delete existing protocol
gui_brainstorm('DeleteProtocol', ProtocolName);
% Create new protocol
gui_brainstorm('CreateProtocol', ProtocolName, 0, 0);
Database requests
The functions bst_get and bst_set allow you to query the database, access the configuration of the software and some display parameters. The complete reference documentation of these functions is included directly in their code (brainstorm3/toolbox/core/bst_get.m and bst_set.m).
Let's start with a few simple examples:
>> ProtocolInfo = bst_get('ProtocolInfo') % Returns the configuration of the current protocol
ProtocolInfo =
Comment: 'TutorialIntroduction'
STUDIES: 'C:\Work\Protocols\TutorialIntroduction\data'
SUBJECTS: 'C:\Work\Protocols\TutorialIntroduction\anat'
iStudy: 6
UseDefaultAnat: 0
UseDefaultChannel: 0
>> isGUI = bst_get('isGUI') % Is the Brainstorm interface currently displayed (0=no, 1=yes)
>> bst_set('FlipYAxis', 1) % The new figures will have the Y axis flipped
>> bst_set('TSDisplayMode', 'butterfly') % The new figures will use a "butterfly" view
>> bst_set('FieldTripDir', FieldTripDir) % Set the path where the FieldTrip toolbox is installedTo reference the files in the database, each protocol is subdivided in Subjects (the "anat" folder and containing the MRI, surfaces and atlases) and Studies (the "data" folder, including the recordings, channel files and all the analyses). Each Study corresponds to a sub-folder (eg. protocol/data/subject01/run01/), and is attached to only one subject.
Subjects and Studies are referenced in the protocol with a unique index most of the time kept in variables named iSubject and iStudy. The files available in them are also referenced with indices, with variables such as iAnatomy, iSurface, iData, iHeadModel, iResults, iTimefreq. You can see the indices in the database explorer by hovering your mouse over the nodes of the tree:

Example: Getting the study structure from the variable sAvgData, defined in the script:
>> sAvgData(1)
ans =
iStudy: 6
iItem: 1
...
>> sStudy = bst_get('Study', sAvgData(1).iStudy) % Get a study structure with its index
sStudy =
Name: 'S01_AEF_20131218_01_600Hz_notch'
FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/brainstormstudy.mat'
DateOfStudy: '13-May-2016'
BrainStormSubject: 'Subject01/brainstormsubject.mat' % Corresponding subject filename
Condition: {'S01_AEF_20131218_01_600Hz_notch'} % Name of the condition (=folder)
Channel: [1x1 struct] % Channel file for the folder
iChannel: [] % Not used anymore
Data: [1x242 struct] % List of "data" files in the folder
HeadModel: [1x1 struct] % List of head models in the folder
iHeadModel: 1 % Default head model (file in green)
Result: [1x244 struct] % List of source files and source links
Stat: [1x0 struct] % List of statistical results
Image: [0x0 struct] % List of images
NoiseCov: [1x2 struct] % Noise(1) and data(2) covariance files
Dipoles: [0x0 struct] % List of dipole files in the folder
Timefreq: [1x247 struct] % List of time-frequency files
Matrix: [0x0 struct] % List of "matrix" files in the folder
% If 1, the trial is marked as bad in the explorerExample: Getting the data structure.
>> sData = sStudy.Data(sAvgData(1).iItem) % Get the structure representing the file from sStudy
sData =
FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat'
Comment: 'Avg: deviant (39 files)' % File comment, displayed in the database explorer
DataType: 'recordings' % Type of data in the file (mostly "recordings")
BadTrial: 0Example: Getting the subject structure.
>> sSubject = bst_get('Subject', sStudy.BrainStormSubject) % Get subject structure from filename
sSubject =
Name: 'Subject01' % Subject name, same as folder name
Comments: '' % Not used much
FileName: 'Subject01/brainstormsubject.mat'
DateOfAcquisition: '' % Not used anymore
Anatomy: [1x1 struct] % List of MRI volumes in the subject folder
Surface: [1x9 struct] % List of surfaces in the subject folder
iAnatomy: 1 % Default MRI volume (in green, index of array Anatomy)
iScalp: 9 % Default head surface (index of array Surface)
iCortex: 4 % Default cortex surface (index of array Surface)
iInnerSkull: [] % Default inner skull surface (index of array Surface)
iOuterSkull: [] % Default outer skull surface (index of array Surface)
iOther: [] % Not used anymore
UseDefaultAnat: 0 % If 1: Subject uses the default anatomy
UseDefaultChannel: 0 % 0=one channel/folder, 1=one/subject, 2=one globalExample: Getting the study structure and data index from a file name.
>> DataFile = sAvgData(1).FileName
DataFile =
Subject01/S01_AEF_20131218_01_600Hz_notch/data_deviant_average_160513_1329.mat
>> [sStudy, iStudy, iData] = bst_get('DataFile', DataFile)
sStudy =
Name: 'S01_AEF_20131218_01_600Hz_notch'
FileName: 'Subject01/S01_AEF_20131218_01_600Hz_notch/brainstormstudy.mat'
DateOfStudy: '13-May-2016'
BrainStormSubject: 'Subject01/brainstormsubject.mat'
Condition: {'S01_AEF_20131218_01_600Hz_notch'}
Channel: [1x1 struct]
iChannel: []
Data: [1x242 struct]
HeadModel: [1x1 struct]
iHeadModel: 1
Result: [1x244 struct]
Stat: [1x0 struct]
Image: [0x0 struct]
NoiseCov: [1x2 struct]
Dipoles: [0x0 struct]
Timefreq: [1x247 struct]
Matrix: [0x0 struct]
iStudy =
6
iData =
1Many other options are available for searching files in the database with bst_get. We cannot list them all in this page, but you can refer to the code of bst_get.m for more information.
To change parameters or database structures: bst_set.m.
File structures
The structures of the different types of files functions were described in the sections "On the hard drive" of the introduction tutorials. Here is a directory of all these sections:
Continuous recordings: File type "raw", Event markers.
Imported recordings: File type "data", Import epochs and Average response.
Power spectrum: File type "timefreq", Power sprectum and frequency filters.
Channel files: File type "channel", Channel file / MEG-MRI coregistration.
SSP/ICA projectors: Saved in the channel files, Artifact cleaning with SSP.
Head models: File type "headmodel", Head model.
Noise covariance: File types "noisecov" and "ndatacov", Noise and data covariance matrices.
Source files: File types "results" and "link", Source estimation.
Time-frequency: File type "timefreq", Time-frequency.
Statistics: File types "pdata", "presults", "ptimefreq" and "pmatrix", Statistics.
MRI volumes: File type "subjectimage", Display the anatomy.
Surfaces: File type "tess", Display the anatomy.
Scouts and atlases: Saved in the surface files, Scouts.
Custom processing
In many situations, you will find useful to read the files available in the database, and maybe modify them. The easiest approaches do not require any scripting, we will start by reviewing them quickly.
Process: Run Matlab command
If you want to modify the values saved in a file (eg. the field "F" from a "data" file), the easiest way is probably to use the process1 File > Run Matlab command. It is also available from Process2 in the category "Other".
It loads the files in input and run them through a piece of Matlab code that you can edit freely. It can extend a lot the flexibility of the Brainstorm pipeline manager, providing an easy access to any Matlab function or script.
The corresponding script looks like this:
sFiles = bst_process('CallProcess', 'process_matlab_eval', sFiles, [], ...
'matlab', 'Data = Data.^2;', ...
'sensortypes', 'MEG', ...
'overwrite', 0);
Export/Import with the database explorer
Right-click on the Deviant average in the database explorer > File > Export to Matlab > "DataMat".
Data exported as "DataMat"
>> DataMat
DataMat =
ChannelFlag: [340x1 double]
ColormapType: []
Comment: 'Avg: deviant (39 files)'
DataType: 'recordings'
Device: 'CTF'
DisplayUnits: []
F: [340x361 double]
History: {45x3 cell}
Std: []
Time: [1x361 double]
nAvg: 39
Events: [1x1 struct]DataMat is the exact content of the corresponding .mat file, as loaded with Matlab's load() function. Edit some of the fields of this structure from the Matlab command window:
>> DataMat.Comment = 'Test square'; >> DataMat.F = DataMat.F .^ 2;
Now right-click on the folder containing the original file > File > Import from Matlab > DataMat:
If instead, you right-click on the original file and select the menu File > Import from Matlab, it overwrites the selected file instead of creating a new one with the selected structure.
Reference: File manipulation
Useful functions for manipulating file names and paths (read the code of the functions for help):
file_fullpath: Converts a relative file path from the Brainstorm database to an absolute path.
file_short: Converts an absolute file path to a short path, relative to the current protocol folder.
file_gettype: Returns the type of a file.
Reading files from a script (all the functions take relative paths in input):
in_bst_data(DataFile): Read an imported epoch.
in_bst_timefreq(TimefreqFile): Read a power spectrum, time-frequency or connectivity file.
in_bst_headmodel(HeadmodelFile, ApplyOrient): Read a head model file and apply orientations.
in_bst_results(ResultsFile, LoadFull): Load a source file and optionally reconstruct the full source time series on the fly (ImagingKernel * recordings).
in_bst_matrix(MatrixFile): Read a file with the type "matrix".
in_bst(FileName, TimeWindow): Read any Brainstorm data file with the possibility to load only a specific part of the file. "TimeWindow" is a range of time values in seconds: [tStart, tStop].
bst_process('LoadInputFile', FileName, Target, TimeWindow): The most high-level function for reading data files. "Target" is a string with the list of signal names or types to load.
bst_memory('GetConnectMatrix', TimefreqMat): Rebuild a full connectivity matrix.
in_fopen(DataFile, FileFormat, ImportOptions): Low-level function for opening continuous files.
in_fread(sFile, ChannelMat, iEpoch, SamplesBounds, iChannels, ImportOptions): Low-level function for reading blocks from continuous files. Require a call to in_fopen first to get the sFile structure.
Saving files:
bst_save(FileName, FileMat, Version, isAppend): Save a file but does not register it in the database.
FileName: Must be an absolute path, use in combination with file_fullpath to use relative paths.
FileMat: Must be a valid Brainstorm structure, corresponding to the file type.
Version: Defines which version of the Matlab .mat format is used to store the data:
- 'v6': Fastest option, bigger files, no compression, no files >2Gb
- 'v7': Slower option, compressed, no files >2Gb
- 'v7.3': Much slower than the others, compressed, but only way to save files > 2Gb.isAppend: If set to 1, updates only the fields defined in FileMat, keep the others untouched.
file_unique: Produces a unique file name by adding a number tag to it.
bst_process('GetNewFilename', OutputFolder, BaseFilename): Generate a new unique file name based on the beginning of the file name (eg. BaseFilename='data_average_test_').
Registering new files in the database:
db_add(iStudy/iSubject, FileMat): Add a new file in an anatomy folder or a functional data folder. This function saves and then registers it in the database. FileMat must be a structure, not a filename. You should not save the file manually before calling this function.
db_add_data(iStudy, FileName, FileMat): Register in the database a structure FileMat that has already been saved in file FileName. You should call bst_save manually before calling this function.
db_add_data(iStudy, FileName, FileMat, iItem): Overwrites the existing file #iItem.
Reload folders (if you saved or deleted files without registering correctly the modification in the database):
db_reload_studies(iStudies): Reload only the select data folders (aka "studies").
db_reload_conditions(iSubjects): Reload all the data folders for a subject.
db_reload_subjects(iSubjects): Reload the anatomy of the selected subjects.
db_reload_database('current'): Reload the entire protocol (anatomy and functional data).
Other useful database functions:
db_add_condition: Create a new folder in a subject.
db_add_subject: Create a new subject.
db_delete_studies: Delete a list of folders.
db_delete_subjects: Delete a list of subjects.
db_group_conditions: Merge two analysis folders from the same subject.
db_rename_condition: Rename a folder.
db_rename_subject: Rename a subject
db_set_channel: Set the channel file for a folder.
db_set_noisecov: Set the noise/data covariance for a folder, or copy to other folders/subjects.
db_set_template: Copy an anatomy template to a subject or use it as the default anatomy.
Export a file from the database to other file formats (read the comments in the functions for help):
- export_channel
- export_data
- export_events
- export_result
- export_timefreq
- export_matrix
- export_mri
- export_surfaces
- export_protocol: Export a subject or an entire protocol as a .zip file.
Convert Brainstorm structures to FieldTrip structures:
- out_fieldtrip_channel
- out_fieldtrip_data
- out_fieldtrip_timefreq
- out_fieldtrip_headmodel
- out_fieldtrip_results
- out_fieldtrip_matrix
Reference: Display functions
Create new visualization figures:
view_channels: Display sensors in a 3D figure.
view_helmet: Display the inner surface of the MEG helmet in a 3D figure.
view_timeseries: Display a data file as time series.
view_timeseries_matrix: Display a custom matrix as time series.
view_topography: Display a data file as a spatial topography.
view_erpimage: Display multiple data files as an image, signal by signal.
view_timefreq: Open a time-frequency file (various display modes available).
view_spectrum: Display power spectrum (PSD or time-frequency files).
view_connect: Open a connectivity matrix (various display modes available).
view_matrix: Open a "matrix" file (various display modes available).
view_contactsheet: Create a contact sheet in time or across a volume from an existing figure.
view_noisecov: Display a noise or data covariance file.
view_dipoles: Open a dipoles file (various display modes available).
view_pac: Open PAC results (various display modes available).
view_mri: View a MRI file in the MRI viewer (with or without a functional overlay from a source file).
view_mri_3d: View a MRI file in a 3D figure (with or without a functional overlay from a source file).
view_surface: View a surface.
view_surface_data: View a surface file with a source file as its texture.
view_surface_matrix: View a custorm surface (user defines vertices, faces, color, transparency).
view_image_reg: Display a 3D or 4D matrix as an image with time and/or frequency dimensions.
view_struct: Display a structure as with the popup menu "File > View file contents".
script_view_sources: Shortcut script to display source files.
bst_memory('UnloadAll', 'Forced'): Close all the existing figures.
Configure time-series figures:
panel_time('SetCurrentTime', t): Set current time.
panel_record('SetTimeLength', duration): Set the length of the current display page, in seconds.
panel_record('SetStartTime', t): Set the start of the current page, in seconds.
panel_record('SetDisplayMode', hFig, DisplayMode): 'column' or 'butterfly'
panel_filter('SetFilters', isLowPass, LowPass, isHighPass, HighPass, isSinRem, SinRem, isMirror)
panel_montage('SetCurrentMontage', hFig, MontageName): Change the montage of channels.
bst_figures('SetSelectedRows', SelectedChannels): Set selected channels (cell array of strings).
figure_timeseries('SetTimeSelectionManual', hFig, TimeWindow): Select a time segment.
Configure 3D figures:
figure_3d('SetStandardView', hFig, 'left'): Change camera view (top,bottom,left,right,front,back).
figure_3d('ViewSensors', hFig, isMarkers, isLabels): Enable the view of sensor markers and labels.
panel_surface('SetShowSulci', hFig, iTess, 1): Enable/Display the display of sulci.
panel_surface('SetSurfaceColor', hFig, iTess, [1 0 0]): Set surface color.
panel_surface('SetSurfaceSmooth', hFig, iTess, Smooth, 0): Set amount of smoothing (0-1).
panel_surface('SetSurfaceTransparency', hFig, iTess, Transp): Set transparency (0-1).
panel_surface('SetDataThreshold', hFig, iTess, Thresh): Set amplitude threshold.
panel_surface('SetSizeThreshold', hFig, iTess, MinSize): Set size threshold (min size slider).
figure_mri('SetLocation', CsName, hFig, [], xyz): CsName=voxel/mri/scs/mni, xyz=[x,y,z]
Configure time-frequency figures:
panel_freq('SetCurrentFreq', iFreq, 1): Set the the current frequency index (from the Freqs vector)
panel_freq('SetCurrentFreq', Freq, 0): Set the the current frequency (in Hz).
sOptions = panel_display('GetDisplayOptions'): Get display options selected in the Display tab.
panel_display('SetDisplayOptions', sOptions): Change the options selected in the Display tab.
sOptions.HideEdgeEffects: Controls the checkbox "Hide edge effects" (0 or 1).
sOptions.HighResolution: Controls the checkbox "Smooth display" (0 or 1).
sOptions.RowName: Controls the signal that is currently displayed (for 'SingleSensor' display mode)
sOptions.Function: Controls the display function (magnitude, power, log, phase).
Configure colormaps:
bst_colormaps('SetColormapName', ColormapType, ColormapName): 'jet', 'parula', 'cmap_rbw', ...
bst_colormaps('SetMaxMode', ColormapType, MaxMode): Colorbar range ('global', 'local', 'custom').
bst_colormaps('SetMaxCustom', ColormapType, [], Min, Max): Set a custom colorbar range.
bst_colormaps('SetColormapAbsolute', ColormapType, isAbsolute): Show positive or relative values
bst_colormaps('SetDisplayColorbar', ColormapType, isDisplay): Show colorbar in the figures.
bst_colormaps('RestoreDefaults', ColormapType): Restore the default configuration for a colormap.
ColormapType: anatomy, meg, eeg, sources, stat1, stat2, time, timefreq, connect1, connectn, image
Configure statistical thresholding:
StatThreshOptions = bst_get('StatThreshOptions'): Get display options selected in the Stat tab.
bst_set('StatThreshOptions', StatThreshOptions): Change the options selected in the Stat tab.
StatThreshOptions.pThreshold: Current p-Value threshold.
StatThreshOptions.Correction: Correction for multiple comparisons ('none', 'fdr', 'bonferroni')
StatThreshOptions.Control: List of dimensions to correct for multiple comparisons (default = [1 2 3])
Export the contents of a figure to a file:
out_figure_image: Screen capture of any Brainstorm figure.
out_figure_movie: Save a movie from one or multitple Brainstorm figures.
out_figure_timefreq: Extract some of the data displayed in a time-frequency figure.
out_figure_timeseries: Extract some of the data displayed in a time series figure.
Example: Creating a new file
This section illustrates how to add new files to the database. We will create a sinusoidal signal and save it in a "matrix" file, in a new folder of subject Test.
% Create two sinsuoidal signals (20Hz,30Hz), over 1 second with a sampling frequency of 1000Hz
t = 0:0.001:1;
F = [sin(20*2*pi*t); 0.5*sin(30*2*pi*t)];
% Initialize an empty "matrix" structure
sMat = db_template('matrixmat');
% Fill the required fields of the structure
sMat.Value = F;
sMat.Comment = 'Test sinusoids';
sMat.Description = {'Signal #1: 20Hz'; 'Signal #2: 30Hz'};
sMat.Time = t;
% Create a new folder "Script" in subject "Test"
iStudy = db_add_condition('Test', 'Script');
% Get the corresponding study structure
sStudy = bst_get('Study', iStudy);There are many options to add a new file to the database, with various levels of requirements. You can call the db_add function (reloads the destination folder, therefore slow if you save many files), save the file in the corresponding folder and reload the protocol, or register the file in the database manually (more complicated but faster).
Option #1: db_add
OutputFile = db_add(iStudy, sMat);
Option #2: bst_save / db_reload_studies
% Another to generate a unique filename (without a timestamp) MatrixFile = file_unique(bst_fullfile(OutputFolder, 'matrix_test.mat')); % Save file bst_save(MatrixFile, sMat, 'v6'); % Reload the folder in which the new file was saved db_reload_studies(iStudy);
Option #3: bst_save / db_add_data
% Get the full path to the new folder (same folder as the brainstormstudy.mat file for this study)
OutputFolder = bst_fileparts(file_fullpath(sStudy.FileName));
% Get a new unique filename (including a timestamp)
MatrixFile = bst_process('GetNewFilename', OutputFolder, 'matrix_test');
% Save file
bst_save(MatrixFile, sMat, 'v6');
% Reference saved file in the database
db_add_data(iStudy, MatrixFile, sMat);
% Update tree
panel_protocols('UpdateNode', 'Study', iStudy);
Example: Editing events
A step that commonly requires manual changes is the definition of the event markers. Many time we have to combine external triggers or behavioral information with the existing events. This example illustrates how to load the events, modify them and save them back.
For the continuous recordings, the events are saved in the .mat file corresponding to the "Link to raw file" available in the database explorer. These structures contain only meta-data and information created with Brainstorm, the EEG/MEG recordings are available in a separate binary file. First, we need to load this link.
% Right-click on a "Link to raw file" in the database explorer > File > Copy file path to clipboard
RawFile = '/.../@rawS01_AEF_20131218_01_600Hz_notch/data_0raw_S01_AEF_20131218_01_600Hz_notch.mat'
% Load the "sFile" structure, contained in the .F structure of the link .mat file
sRaw = in_bst_data(RawFile, 'F');
>> sRaw.F.events
ans =
1x7 struct array with fields:
label
color
epochs
samples
times
reactTimes
selectFor example, let's say we want to add 30ms to all the events in the category "button", to compensate for some hardware delay, and create a new event category with the modified timing. We need first to identify what is the index of the category "button", in this array of 7 events structures.
% Find the index of the event category "button"
iEvtButton = find(strcmpi({sRaw.F.events.label}, 'button'));
>> iEvtButton
iEvtButton =
3In the code above, note this special Matlab syntax that allows the concatenation of the values of one field across multiple structures, in an array of structures:
>> {sRaw.F.events.label}
ans =
'standard' 'deviant' 'button' 'cardiac' 'blink' 'bad_1-7Hz' 'bad_40-240Hz'If you wanted to search instead all the events containing a specific tag, for example "bad", you can use the cellfun function (function that applies the same function sequentially to all the elements in a cell array and concatenates the results) in combination with the strfind function (search for a substring). The final call to the find function returns at which indices the list of tags found in the event label is not empty.
>> iEvtBad = find(~cellfun(@(c)isempty(strfind(c,'bad')), {sRaw.F.events.label}))
iEvtBad =
6 7The code below copies the existing event category "button", renames it and add a 30ms offset. Note that times and samples field must always match. If you modify one, you must also modify the others (times = samples/sfreq). If you add or remove events, you must adjust the size of the epochs field (always 1 for most file formats).
% Copy the event category "button" to a new cagory
iEvtNew = length(sRaw.F.events) + 1;
sRaw.F.events(iEvtNew) = sRaw.F.events(iEvtButton);
% Rename new event to "button_offset"
sRaw.F.events(iEvtNew).label = 'button_offset';
% Compute how many samples correspond to 30ms (18 samples at 600Hz = 30ms)
offsetSample = round(0.030 .* sRaw.F.prop.sfreq);
% Apply this offset to the events in the "button_offset" category
sRaw.F.events(iEvtNew).samples = sRaw.F.events(iEvtNew).samples + offsetSample;
% Re-compute the corresponding times
sRaw.F.events(iEvtNew).times = sRaw.F.events(iEvtNew).samples ./ sRaw.F.prop.sfreq;
% Re-generate an epochs field with only ones (optional here, as we didn't change the number of evt)
sRaw.F.events(iEvtNew).epochs = ones(1, size(sRaw.F.events(iEvtNew).times, 2));
% Change the event color to yellow (red=1, green=1, blue=0)
sRaw.F.events(iEvtNew).color = [1 1 0];
>> sRaw.F.events(iEvtNew)
ans =
label: 'button_offset'
color: [1 1 0]
epochs: [1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1]
samples: [1x40 double]
times: [1x40 double]
reactTimes: []
select: 1The last step is to save the modifications back to the "Link to raw file". Here the call to file_fullpath is optional because the variable RawFile was already containing the absolute path to the file.
% Update the sRaw structure to the RawFile file (the last parameter appends to the existing struct) bst_save(file_fullpath(RawFile), sRaw, 'v6', 1);
Open the recordings to make sure your transformation worked the way you expected.
Find examples in the code
The easier way to understand how to use a function is to search the code with the "Find files" interface in Matlab. Go to the brainstorm3 folder, click on "File files" (or Ctrl+Shift+F), enter the name of function your are looking for in "Find files containing text", Include subfolders, Match case. It will return all the lines that include the string you entered across all the files in Brainstorm. Just double-click on a line to jump to the code in the Matlab editor.
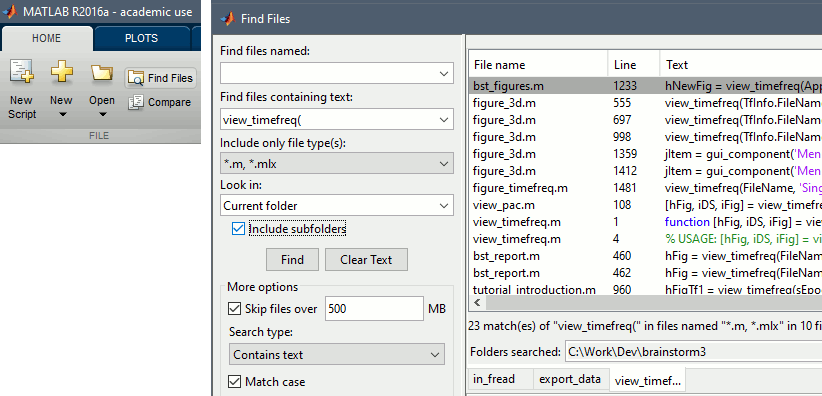
You can use the same interface to find what function is called when you click on a button or menu in the interface. Search for the label or the tooltip of the interface element in the same way. The example below shows how to track what happens when you click on the headmodel popup menu "Check spheres".
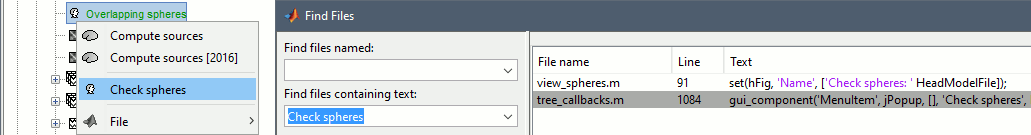
If you have trouble understanding how to set some input parameters, you can use the debugger to explore a real use case. Place a breakpoint at the begging of your function of interest (watch this tutorial if you don't know how to do it), for in example view_timeseries.m. Then click on the corresponding menus in the Brainstorm interface (eg. double-click on on a data file). When the execution reaches the line you selected, it stops and give you back the commands. You can explore the values in all the variables, modify them, and execute the code step by step (many options available in the Editor tab of Matlab).
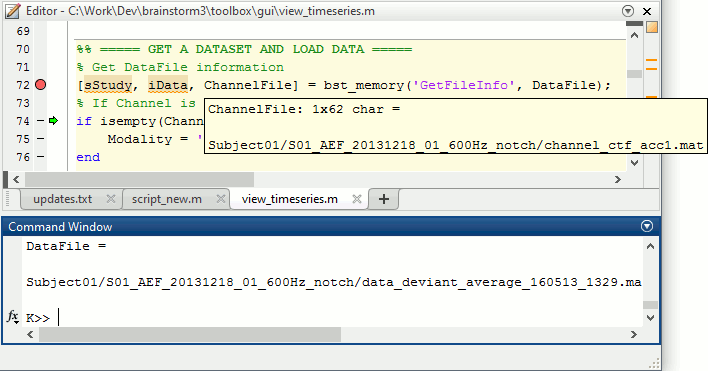
Additional quality control [TODO]
You cann add in the reports all the information that will help you control the quality of the analysis, or even figures you want to include in publications or clinical reports.
Create figures and snapshot them
% Display raster plot
hFigRaster = view_erpimage({sFilesEpochs1.FileName}, 'erpimage', 'MEG');
bst_report('Snapshot', hFigRaster, sFilesEpochs1(1).FileName, 'ERP image');Add messages: warnings and errors:
Loop over subject and runs [TODO]
Creating loops is not supported yet by the script generator, but relatively easy to do from a script without having to know too much about Matlab programming.
1) Fill the cell array SubjectNames with all your subjects names, with the same dimensions as the list of input raw files (sFiles)
- 2) Add a "for" loop that includes all the bst_process() calls (leave the bst_report() calls and input definition outside)
3) Inside the loop, replace SubjectNames with SubjectNames{i} and sFiles with sFiles(i)
How to process an entire study
This section proposes a standard workflow for processing a full group study with Brainstorm. It contains the same steps of analysis as the introduction tutorials, but separating what can be done automatically from what should be done manually. This workflow can be adapted to most ERP studies (stimulus-based).
Prototype: Start by processing one or two subjects completely interactively (exactly like in the introduction tutorials). Use the few pilot subjects that you have for your study to prototype the analysis pipeline and check manually all the intermediate stages. Take notes of what you're doing along the way, so that you can later write a script that reproduces the same operations.
Anatomical fiducials: Set NAS/LPA/RPA and compute the MNI transformation for each subject.
Segmentation: Run FreeSurfer/BrainSuite to get surfaces and atlases for all the subjects.
File > Batch MRI fiducials: This menu prompts for the selection of the fiducials for all the subjects and saves a file fiducials.m in each segmentation folder. You will not have to redo this even if you have to start over your analysis from the beginning.
Script: Write a loop that calls the process "Import anatomy folder" for all the subjects.
Alternatives: Create and import the subjects one by one and set the fiducials at the import time. Or use the default anatomy for all the subjects (or use warped templates).
Script #1: Pre-processing: Loop on the subjects and the acquisition runs.
Create link to raw files: Link the subject and noise recordings to the database.
Event markers: Read and group triggers from digital and analog channel, fix stimulation delays
Evaluation: Power spectrum density of the recordings to evaluate their quality.
Pre-processing: Notch filter, sinusoid removal, band-pass filter.
Evaluation: Power spectrum density of the recordings to make sure the filters worked well.
Cleanup: Delete the links to the original files (the filtered ones are copied in the database).
Detect artifacts: Detect heartbeats, Detect eye blinks, Remove simultaneous.
Compute SSP: Heartbeats, Blinks (this selects the first component of each decomposition)
Compute ICA: If you have some artifacts you'd like to remove with ICA (no default selection).
Screenshots: Check the MRI/sensors registration, PSD before and after corrections, SSP.
Export the report: One report per subject, or one report for all the subjects, saved in HTML.
Manual inspection #1:
Check the reports: Information messages (number of events, errors and warnings) and screen captures (registration problems, obvious noisy channels, incorrect SSP topographies).
Mark bad channels: Open the recordings, select the channels and mark them as bad. Or use the process "Set bad channels" to mark the same bad channels in multiple files.
Fix the SSP/ICA: For the suspicious runs: Open the file, adjust the list of blink and cardiac events, remove and recompute the SSP decompositions, manually select the components.
Detect other artifacts: Run the process on all the runs of all the subjects at once (select all the files in Process1 and run the process, or generate the equivalent script).
Mark bad segments: Review the artifacts detected in 1-7Hz and 40-240Hz, keep only the ones you really want to remove, then mark the event categories as bad. Review quickly the rest of the file and check that there are no other important artifacts.
Additional SSP: If you find one type of artifact that repeats (typically saccades and SQUID jumps), you can create additional SSP projectors, either with the process "SSP: Generic" or directly from a topography figure (right-click on the figure > Snapshot> Use as SSP projector).
Script #2: Subject-level analysis: Epoching, averaging, sources, time-frequency.
Importing: Process "Import MEG/EEG: Events" and "Pre-process > Remove DC offset".
Averaging: Average trials by run, average runs by subject (registration problem in MEG).
Noise covariance: Compute from empty room or resting recordings, copy to other folders.
Head model: Compute for each run, or compute once and copy if the runs are co-registered.
Sources: Compute for each run, average across runs and subjects in source space for MEG.
Time-frequency: Computation with Hilbert transform or Morlet wavelets, then normalize.
Screenshots: Check the quality of all the averages (time series, topographies, sources).
Export the report: One report per subject, or one report for all the subjects, saved in HTML.
Manual inspection #2:
Check the reports: Check the number of epochs imported and averaged in each condition, check the screen capture of the averages (all the primary responses should be clearly visible).
Regions of interest: If not using predefined regions from an atlas, define the scouts on the anatomy of each subject (or on the template and then project them to the subjects).
Script #3: Group analysis, ROI-based analysis, etc.
Averaging: Group averages for the sensor data, the sources and the time-frequency maps.
Statistics: Contrast between conditions or groups of subjects.
Regions of interest: Any operation that involve scouts.
Final script
The following script from the Brainstorm distribution reproduces the introduction tutorials ("Get started"): brainstorm3/toolbox/script/tutorial_introduction.m - Report: report_TutorialIntroduction.html
For an example of a script illustrating how to create loops, look at the tutorial MEG visual: single subject. brainstorm3/toolbox/script/tutorial_visual_single.m - Report: report_TutorialVisual_sub001.html
Additional documentation
Tutorial: How to write your own process