|
Size: 2982
Comment:
|
Size: 3065
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 58: | Line 58: |
| * Then place the cross on the anterior commissure ([[CoordinateSystems|see this page if our in trouble]]) | * Then place the cross on the anterior commissure ([[CoordinateSystems|see this page if you are in trouble]]) |
| Line 61: | Line 61: |
| * Uncheck the "Cortical Fold Graph" step, we don't need that. | |
| Line 68: | Line 69: |
| What if it cr |
T1-MRI Segmentation with BrainVISA
It is not the purpose of Brainstorm tutorials to teach you how to use BrainVISA. But most Brainstorm users are lost when it reaches to the segmentation of the MRI. So here is a short introduction of the BrainVISA T1 Pipeline.
To extract head and cortex meshes from a T1 MRI, you can also try to use: BrainSuite or ?FreeSurfer.
Installation
Download the latest version of BrainVISA from http://brain.info.
Install it on your computer by following the instructions on the download page.
- Start BrainVISA
You'll have to create a database for saving your files: this will be either asked at startup, or you'll have to !BrainVISA menu > Preferences:
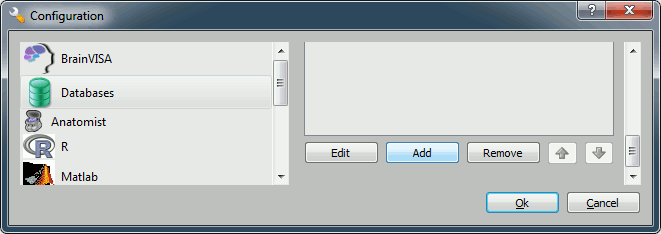
Click on Add, and create a directory with is not in any Brainstorm directory. For example:
Windows: My Documents\brainvisa_db\
Linux: /home/username/brainvisa_db/
- MacOS: Documents/brainvisa_db/
- Click on Ok.
- Your are ready to import and process your MRI.
Import MRI
To process a MRI volume with BrainVISA, you have first to import it in the database.
Select the process: T1-MR1 > Import > Import T1 MRI, and click on Open button.
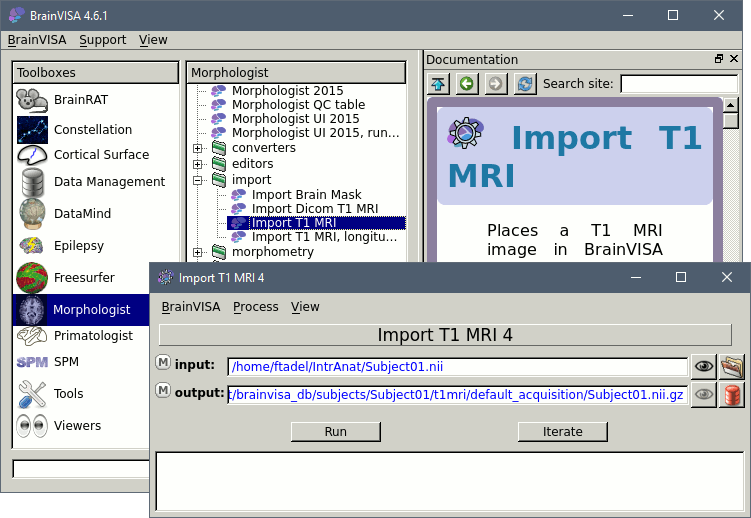
The following figure will appear:
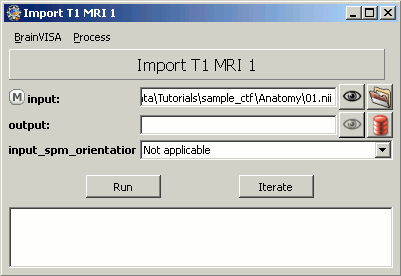
As an example, you can take the MRI from the 4D-Neuroimaging tutorial (bst_sample_4d.zip accessible from Download section).
Select the input file.
To select the output file: Click on the red button, fill the following fields:
- Protocol: segmentation
- Subject: 4D
- A default filename should appear, and you just have to click on Ok.
Click on Run, wait for the process to end. Done. You can close this window.
Configure T1 pipeline
Back to BrainVISA window, now select process T1 MRI > Segmentation Pipeline > T1 Pipeline.
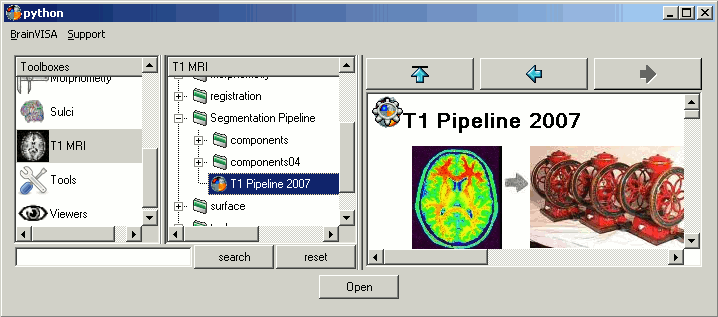
Click on open.
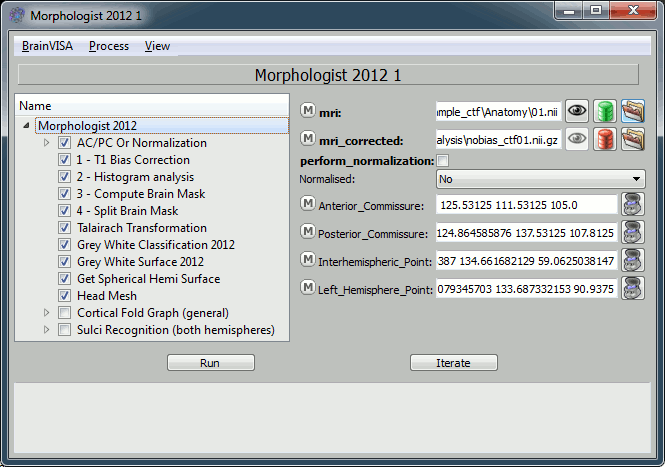
How to fill all those fields:
- Mri: click on the green button, and select from the database the MRI you've just imported
- Anterior commissure:
- Click on the "Anatomist" button. This will start anatomist.
- In Anatomist window, look for a view that gives you an axial view
Then place the cross on the anterior commissure (see this page if you are in trouble)
- The go back to the "T1 Pipeline" window again, and click again on "Anatomist" button. This should write the current position of the cursor in the "Anterior commissure" field.
- Repeat for all the other points: Posterior commissure, Interhemispheric point, Left hemisphere point
- Uncheck the "Cortical Fold Graph" step, we don't need that.
The go to the "AC/PC Or Normalization" step, and select true for the option allow_flip_initial_MRI.
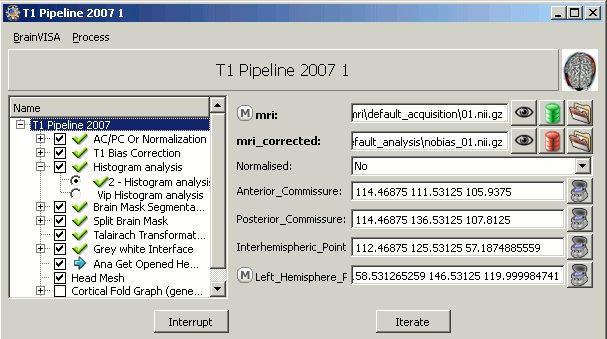
Click on Run and pray hard.
What if it cr
