|
Size: 13234
Comment:
|
← Revision 83 as of 2022-06-24 10:19:19 ⇥
Size: 14441
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| It is not the purpose of Brainstorm tutorials to teach you how to use BrainVISA. But many Brainstorm users are lost when it gets to the segmentation of the MRI. So here is a short introduction to the BrainVISA T1 MRI processing pipeline. To extract head and cortex meshes from a T1 MRI, you can also try to use: [[http://www.loni.ucla.edu/Software/BrainSuite|BrainSuite]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. | ''Authors: Francois Tadel'' |
| Line 4: | Line 4: |
| We are going to illustrate the use of BrainVISA with the MRI from the CTF tutorials. You should already have those files on your computer, if you followed the basic tutorials. If it is not the case, go to the [[http://neuroimage.usc.edu/brainstorm3_register/download.php|Download page]], and get the file sample_ctf.zip. | It is not the purpose of Brainstorm tutorials to teach you how to use BrainVISA. But many Brainstorm users are lost when it gets to the segmentation of the MRI. So here is a short introduction to the BrainVISA T1 MRI processing pipeline. To extract head and cortex meshes from a T1 MRI, you can also try to use: [[Tutorials/SegBrainSuite|BrainSuite]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. |
| Line 6: | Line 6: |
| This tutorial was written for BrainVISA 4.3. To get started, or for additional information, you can also read the [[http://brainvisa.info/doc/axon/bv_training/en/html/index.html|BrainVISA tutorial pages]]. | This tutorial was written for BrainVISA 4.6. To get started, or for additional information, you can also read the [[http://www.brainvisa.info/axon-4.6/user_doc/tutorial.html|BrainVISA tutorial]] and the [[http://brainvisa.info/axon-4.6/user_doc/morphologist_tutorial.html|Morphologist tutorial]]. <<TableOfContents(2,2)>> |
| Line 10: | Line 12: |
| * Install it on your computer by following the instructions on the [[http://brainvisa.info/download.html#installation|download page]]. | * Install it on your computer by following the instructions on the [[http://brainvisa.info/web/download.html|download page]]. |
| Line 22: | Line 24: |
| == Running BrainVISA == | == Running Morphologist == The procedure to run the segmentation of a MRI volume is described in the [[http://brainvisa.info/axon-4.6/user_doc/morphologist_tutorial.html|Morphologist tutorial]]. Only the first few steps are illustrated below. ==== Import the MRI ==== * Double-click on the process: Morphologist > import > Import T1 MRI. * In the first option line "input", click on the button that says "Browse the filesystem (load mode)", the last one on the line. Select the MRI file you want to process. * In the second line "output", you can click on the red button "Browse the database (save mode)" to define where and how this MRI file should be imported in the database. You can edit the subject name and other properties. The click on the button "Run" to import the file.<<BR>><<BR>> {{attachment:importMri.gif}} |
| Line 24: | Line 33: |
| The T1 MRI processing pipeline in BrainVISA is now called Morphologist. Double-click on the icon "Morphologist 2012" to get started. {{attachment:morphologist.gif}} The Morphologist window shows on the left the list of analysis steps that are part of this T1 MRI processing pipeline. They can be selected or unselected independently. When you click on a step, it shows all the possible options and input and output files on the right. {{attachment:morphologist2.gif|morphologist.gif}} ==== Select the MRI file ==== The only options than we need to set (hopefully), are the global ones, that you get when you click on the top element in the list (Morphologist 2012). Let's start with the selection of the MRI file: * On the "mri" line, click on the button that says "Browse the filesystem (load mode)", the last one on the line, that represents a folder. The other button, the green one, is for selecting a subject that is already in the database. * Select the tutorial file sample_ctf/Anatomy/'''01.nii''' <<BR>><<BR>> {{attachment:selectFile.gif|morphologist.gif}} * On the "mri_corrected" line, click on the red button that says "Browse the database (save mode)", the second to last one. Enter the following options:<<BR>><<BR>> {{attachment:output.gif}} <<BR>><<BR>> * File format: NIfTI-1 image * Protocol: segmentation * Subject: ctf01 * Click on ok. ==== Setting the options ==== * '''Anterior commissure''': * Click on the "Anatomist" button (at the end of the line). This will start anatomist. * In Anatomist window, look for a view that gives you an axial view * Then place the cross on the anterior commissure (see page: CoordinateSystems) * Go back to the "T1 Pipeline" window again, and click again on "Anatomist" button for the anterior commissure. This should write the current position of the cursor in the "Anterior commissure" field. <<BR>><<BR>> {{attachment:anatomist.gif|output.gif}} <<BR>><<BR>> * '''Posterior commissure''': Same operation * '''Interhemispheric point''': Same operation, pick it on the top part of the brain. * '''Left hemisphere point''': Same, but be VERY carful with this. By default, contrary to Brainstorm, the view in Anatomist is in radiological convention. Which means that the left point you have to pick is shown '''on the right part on the axial slice'''. * If for some reason the MRI you are importing has the left and right flipped, you should click on the left part of the axial slice, and then set the option "allow_flip_initial_MRI" to "true" in the "AC/PC step". * Keep all the other options to their default values. * Just '''uncheck '''the steps: '''Cortical Fold Graph''' and '''Sulci recognition'''. We don't need those two, and they take a lot of time. * Click on Run and pray hard. It doesn't take long, but it may crash.<<BR>><<BR>> {{attachment:T1PipelinePrepare.gif|output.gif}} |
* Double-click on the process "Morphologist 2015" to get started.<<BR>><<BR>> {{attachment:morphologist.gif}} * The Morphologist window shows on the left the list of analysis steps that are part of this T1 MRI processing pipeline. They can be selected or unselected independently. When you click on a step, it shows all the possible options and input and output files on the right. * The only options than we need to set (hopefully), are the global ones, that you get when you click on the top element in the list (Morphologist 2015). Let's start with the selection of the MRI file. * On the "t1mri" line, click on the green button "Browse the database (load mode)". Select the subject you just imported and click on "Ok" to validate. All the other file names in the options should get filled automatically by default paths.<<BR>><<BR>> {{attachment:selectMri.gif}} <<BR>><<BR>> {{attachment:morphologist2.gif|morphologist.gif}} * If you have Matlab and SPM12 installed, you can use the default normalization method (as illustrated in the screen capture above). Otherwise, you should select the option "Prepare subject for analysis pipeline" and place the AC/PC/IH referential points manually. * If you would like to obtain the [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegBrainVisa#Cortical_parcellations|MarsAtlas parcellation]] for this subject: * Select the checkbox '''Left Hemisphere''' > '''Sulci Recognition''' * Select the checkbox '''Right Hemisphere''' > '''Sulci Recognition''' * Select the checkbox '''Sulci graph morphometry by subject''' * Additional or updated instructions on the BrainVISA website: <<BR>>http://brainvisa.info/axon-4.6/en/processes/HipHopCorticalParameterization.html * Click on '''Run''' to start the Morphologist segmentation pipeline. |
| Line 58: | Line 46: |
| * Try changing the options of the process that crashed * Post your problem on the BrainVISA forum * Try another software, such as [[Tutorials/LabelFreeSurfer|FreeSurfer]] |
* Try changing the options of the process that crashed. * Post your problem on the [[http://brainvisa.info/forum/|BrainVISA forum]]. * Try another software, such as [[Tutorials/LabelFreeSurfer|FreeSurfer]]. |
| Line 62: | Line 50: |
| <<BR>><<BR>> | ==== MarsAtlas parcellation ==== * In BrainVISA, select the process:<<BR>> '''Cortical surface''' > '''Anatomy''' > '''Hip-Hop Cortical Parameterization''' * Keep all the options selected and click on Run to start the computation.<<BR>><<BR>> {{attachment:marsatlas_compute.gif||width="697",height="467"}} |
| Line 67: | Line 57: |
| 1. Right-click on the subject > Import BrainVISA folder...<<BR>><<BR>> {{attachment:import1.gif}} <<BR>><<BR>> 1. Select the top folder of your subject: /.../brainvisa_db/segmentation/ctf01 |
1. Right-click on the subject > Import anatomy folder...<<BR>><<BR>> {{attachment:import1.gif}} <<BR>><<BR>> 1. Select the file format "BrainVISA folder" and select the top folder of your subject: /.../brainvisa_db/segmentation/SubjectName |
| Line 70: | Line 60: |
| 1. The MRI Viewer appears, and a help window asks you to validate the orientation of the MRI and to define the 3 missing fiducial points (NAS/LPA/RPA, as the three others are already imported). If something doesn't look right at this step, for instance if the MRI is not presented with a correct orientation, you should stop this automatic import process and follow the manual instructions in the basic tutorial pages. <<BR>><<BR>> {{attachment:mriviewer.gif}} <<BR>><<BR>> 1. Place the three fiducials NAS/LPA/RPA. If you need help, refer to this page: CoordinateSystems |
1. The MRI Viewer appears. The AC/PC/IH points are already read from the BrainVISA folder, but you should define the positions of the NAS/LPA/RPA [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems|fiducial points]]. Alternatively, compute the [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|MNI normalization]] for obtaining default positions for these points.<<BR>><<BR>> {{attachment:mriviewer.gif}} <<BR>><<BR>> |
| Line 73: | Line 62: |
| 1. This process imports the following files from the folder ...'''/ctf01/t1mri/default_acquisition/default_analysis/''' :<<BR>> * '''nobias_ctf01.nii.gz''' (T1 MRI volume) * '''ctf01.APC''' (position of the fiducials AC, PC and IH) * segmentation/mesh/'''ctf01_head.gii''' (head surface) * segmentation/mesh/'''ctf01_Lhemi.gii''' (grey/csf interface, left hemisphere) * segmentation/mesh/'''ctf01_Rhemi.gii''' (grey/csf interface, right hemisphere) * segmentation/mesh/'''ctf01_Lwhite.gii''' (white matter, left hemisphere) * segmentation/mesh/'''ctf01_Rwhite.gii''' (white matter, right hemisphere) 1. The successive steps that are performed automatically by Brainstorm: |
1. This process imports the following files from the folder ...'''/SubjectName/t1mri/default_acquisition/default_analysis/''' :<<BR>> * '''nobias_SubjectName.nii.gz''' (T1 MRI volume) * SubjectName'''.APC''' (position of the fiducials AC, PC and IH) * segmentation/mesh/'''SubjectName_head.gii''' (head surface) * segmentation/mesh/'''SubjectName_Lhemi.gii''' (grey/csf interface, left hemisphere) * segmentation/mesh/'''SubjectName_Rhemi.gii''' (grey/csf interface, right hemisphere) * segmentation/mesh/'''SubjectName_Lwhite.gii''' (white matter, left hemisphere) * segmentation/mesh/'''SubjectName_Rwhite.gii''' (white matter, right hemisphere) * segmentation/mesh/surface_analysis/SubjectName'''_Lwhite_parcels_marsAtlas.gii''' * segmentation/mesh/surface_analysis/SubjectName'''_Rwhite_parcels_marsAtlas.gii''' 1. The successive steps that are automatically performed by Brainstorm: |
| Line 83: | Line 74: |
| * Import the MarsAtlas parcellation for the white surfaces | |
| Line 89: | Line 81: |
| * '''ctf01_head_8000V''': Scalp surface generated by Brainvisa (holes filled by Brainstorm). * '''cortex_70000V''': High-resolution cortex surface that was generated by BrainVISA.This one appears in green, it means that is going to be used as the default by the processes that require a cortex surface. * '''cortex_15000V''': Low-resolution cortex surface, downsampled using the '''reducepatch''' function from Matlab (it keeps a meaningful subset of vertices from the original surface). * '''white_70000V''': High-resolution white matter envelope from BrainVISA * '''white_15000V''': Low-resolution white matter, processed with reducepatch 1. A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly generated and imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.<<BR>><<BR>> |
* '''SubjectName_head_8000V''': Scalp surface generated by Brainvisa (holes filled by Brainstorm). * '''cortex_170000V''': High-resolution cortex surface that was generated by BrainVISA. * '''cortex_15000V''': Low-resolution cortex surface, downsampled using the '''reducepatch''' function from Matlab (it keeps a meaningful subset of vertices from the original surface). This surface appears in green, it means that is going to be used as the default by the processes that require a cortex surface. * '''white_170000V''': High-resolution white matter surface from BrainVISA (with MarsAtlas) * '''white_15000V''': Low-resolution white matter, processed with reducepatch (with MarsAtlas) 1. A figure is automatically shown at the end of the process, in order to check visually that the low-resolution cortex and head surfaces were properly imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.<<BR>><<BR>> |
| Line 109: | Line 101: |
| * Ask help from the developer through the BrainVISA user forum | * Ask help from the developer through the [[http://brainvisa.info/forum/|BrainVISA user forum]] |
| Line 115: | Line 107: |
| The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can align automatically the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, those points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures. | The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can automatically align the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, these points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures. |
| Line 117: | Line 109: |
| {{attachment:checkAlignMeg.gif||height="243",width="298"}} | {{attachment:checkAlignMeg.gif||width="298",height="243"}} |
| Line 125: | Line 117: |
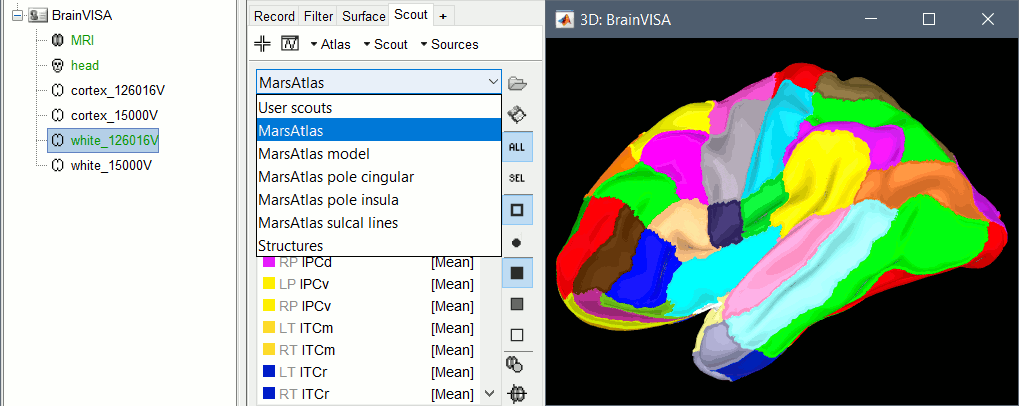
| You can also try to import manually the original BrainVISA head surface. Right click on the subject > Import surface > ctf01_head_8000V. If the surface has holes, you can right-click on the head surface > Fill holes, and then use a different value than the one used by default in the automatic procedure (2). | == Cortical parcellations == The process '''Cortical surface''' > '''Anatomy''' > '''Hip-Hop Cortical Parameterization''' in BrainVISA implements an automatic parcellation of the cortical surface in anatomical regions. The '''MarsAtlas''' model is described here:<<BR>>https://meca-brain.org/software/marsatlas/ This atlas is available only on the white matter surface (ie. the grey-white interface, named white_*). It is imported in Brainstorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution white surface > File > View file contents. You can see the available structures "Atlas", the first one that has Name='User scouts', and the second one Name='MarsAtlas'. {{attachment:viewMat.gif||width="386",height="470"}} To access them from the interface: Double-click on the cortex and go to the ''Scout'' tab, and click on the drop-down list to select another ''Atlas ''(ie group of scouts): {{attachment:marsAtlas.gif||width="656",height="261"}} |
| Line 128: | Line 129: |
| In case you need to import the MRI, surfaces and atlases separately instead of using the menu "Import BrainVISA folder", here is the sequence of operations to perform to get to the same result: | In case you need to import the MRI, surfaces and atlases separately instead of using the menu "Import anatomy folder", here is the sequence of operations to perform to get to the same result: |
| Line 132: | Line 133: |
| 1. Set the 6 fiducial points, save | 1. Set the 3 fiducial points NAS/LPA/RPA (or compute MNI normalization), save |
| Line 134: | Line 135: |
| 1. Double-click on Lwhite to display it. In the scout tab: Atlas > Load atlas > select the file format "GIfTI texture" and the file default_analysis/segmentation/mesh/surface_analysis/..._Lwhite_parcels_marsAtlas.gii. Close the figure. 1. Repeat with surface Rwhite. |
|
| Line 142: | Line 145: |
| == Feedback == <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/SegBrainVisa)>> |
<<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/SegBrainVisa)>> |
Using BrainVISA
Authors: Francois Tadel
It is not the purpose of Brainstorm tutorials to teach you how to use BrainVISA. But many Brainstorm users are lost when it gets to the segmentation of the MRI. So here is a short introduction to the BrainVISA T1 MRI processing pipeline. To extract head and cortex meshes from a T1 MRI, you can also try to use: BrainSuite, CAT12 or FreeSurfer.
This tutorial was written for BrainVISA 4.6. To get started, or for additional information, you can also read the BrainVISA tutorial and the Morphologist tutorial.
Contents
Installation
Download the latest version of BrainVISA from http://brainvisa.info
Install it on your computer by following the instructions on the download page.
- Start BrainVISA
You have to create a database for storing your files: this will be either asked at startup, or you'll have to select the menu BrainVISA > Preferences:
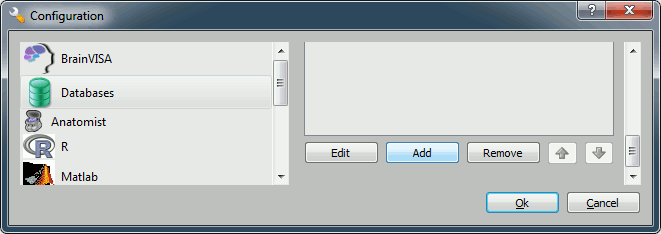
Click on Add, and create a directory with is not in any Brainstorm directory. For example:
Windows: My Documents\brainvisa_db\
Linux: /home/username/brainvisa_db/
MacOS: Documents/brainvisa_db/
- Click on Ok.
- Your are ready to import and process your MRI.
Running Morphologist
The procedure to run the segmentation of a MRI volume is described in the Morphologist tutorial. Only the first few steps are illustrated below.
Import the MRI
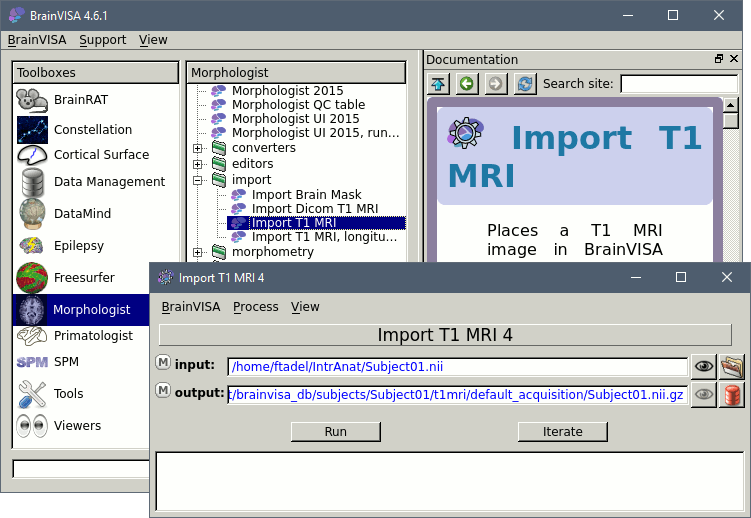
Double-click on the process: Morphologist > import > Import T1 MRI.
- In the first option line "input", click on the button that says "Browse the filesystem (load mode)", the last one on the line. Select the MRI file you want to process.
In the second line "output", you can click on the red button "Browse the database (save mode)" to define where and how this MRI file should be imported in the database. You can edit the subject name and other properties. The click on the button "Run" to import the file.
Start Morphologist
Double-click on the process "Morphologist 2015" to get started.
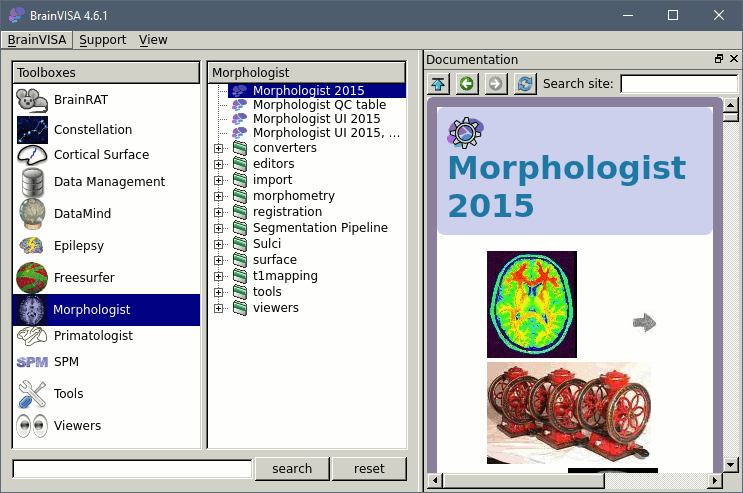
- The Morphologist window shows on the left the list of analysis steps that are part of this T1 MRI processing pipeline. They can be selected or unselected independently. When you click on a step, it shows all the possible options and input and output files on the right.
- The only options than we need to set (hopefully), are the global ones, that you get when you click on the top element in the list (Morphologist 2015). Let's start with the selection of the MRI file.
On the "t1mri" line, click on the green button "Browse the database (load mode)". Select the subject you just imported and click on "Ok" to validate. All the other file names in the options should get filled automatically by default paths.

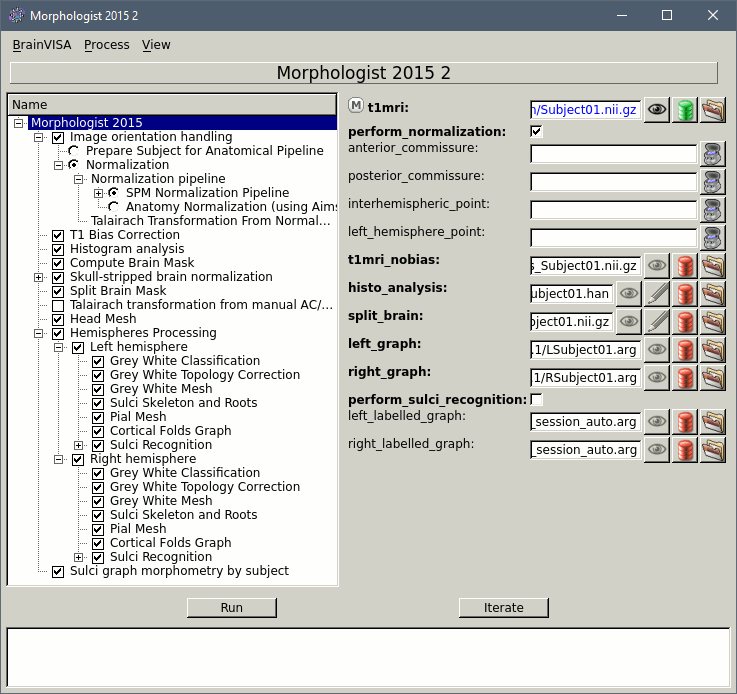
- If you have Matlab and SPM12 installed, you can use the default normalization method (as illustrated in the screen capture above). Otherwise, you should select the option "Prepare subject for analysis pipeline" and place the AC/PC/IH referential points manually.
If you would like to obtain the MarsAtlas parcellation for this subject:
Select the checkbox Left Hemisphere > Sulci Recognition
Select the checkbox Right Hemisphere > Sulci Recognition
Select the checkbox Sulci graph morphometry by subject
Additional or updated instructions on the BrainVISA website:
http://brainvisa.info/axon-4.6/en/processes/HipHopCorticalParameterization.html
Click on Run to start the Morphologist segmentation pipeline.
What if if crashes?
- Try changing the options of the process that crashed.
Post your problem on the BrainVISA forum.
Try another software, such as FreeSurfer.
MarsAtlas parcellation
In BrainVISA, select the process:
Cortical surface > Anatomy > Hip-Hop Cortical ParameterizationKeep all the options selected and click on Run to start the computation.

Importing the results in Brainstorm
- Switch to the anatomy side of the database explorer
- Create a new subject, set the default anatomy option to "No, use individual anatomy"
Right-click on the subject > Import anatomy folder...
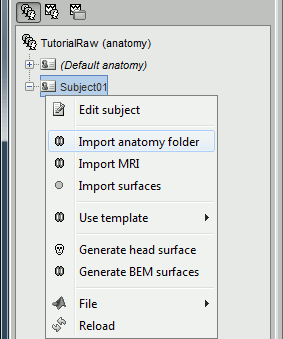
- Select the file format "BrainVISA folder" and select the top folder of your subject: /.../brainvisa_db/segmentation/SubjectName
Then you're prompted for the number of vertices you want in the final cortex surface. This will by extension define the number of dipoles to estimate during the source estimation process. By default we set this value to 15000 for the entire brain (it means 7500 for each hemisphere).
The MRI Viewer appears. The AC/PC/IH points are already read from the BrainVISA folder, but you should define the positions of the NAS/LPA/RPA fiducial points. Alternatively, compute the MNI normalization for obtaining default positions for these points.
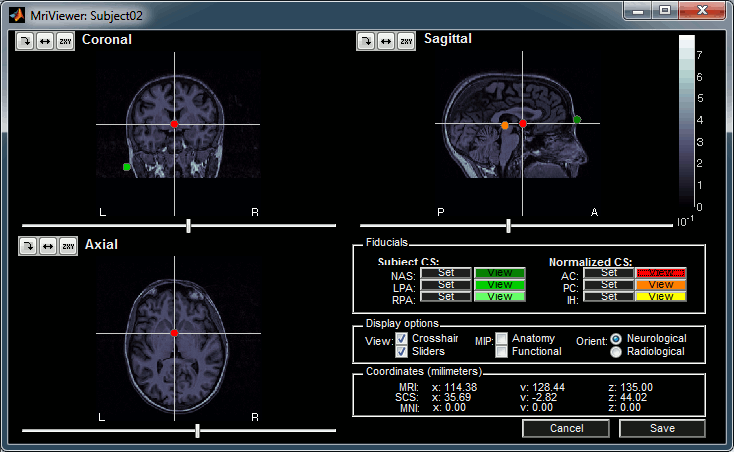
- Click on Save to keep your modifications, and the automatic import will go on.
This process imports the following files from the folder .../SubjectName/t1mri/default_acquisition/default_analysis/ :
nobias_SubjectName.nii.gz (T1 MRI volume)
SubjectName.APC (position of the fiducials AC, PC and IH)
segmentation/mesh/SubjectName_head.gii (head surface)
segmentation/mesh/SubjectName_Lhemi.gii (grey/csf interface, left hemisphere)
segmentation/mesh/SubjectName_Rhemi.gii (grey/csf interface, right hemisphere)
segmentation/mesh/SubjectName_Lwhite.gii (white matter, left hemisphere)
segmentation/mesh/SubjectName_Rwhite.gii (white matter, right hemisphere)
segmentation/mesh/surface_analysis/SubjectName_Lwhite_parcels_marsAtlas.gii
segmentation/mesh/surface_analysis/SubjectName_Rwhite_parcels_marsAtlas.gii
- The successive steps that are automatically performed by Brainstorm:
- Import all the surfaces (left/right, white/cortex)
Import the MarsAtlas parcellation for the white surfaces
- Downsample each hemisphere to the number specified in the options (by default 7500, half of the total default number 15000)
- Merge left and right hemispheres for the two surface types: white matter and cortex envelope
- Delete all the unnecessary surfaces
- Fill the holes in the head surface
The files you can see in the database explorer at the end:
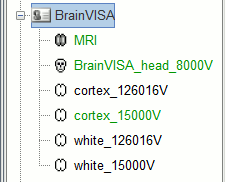
MRI: The T1 MRI of the subject
SubjectName_head_8000V: Scalp surface generated by Brainvisa (holes filled by Brainstorm).
cortex_170000V: High-resolution cortex surface that was generated by BrainVISA.
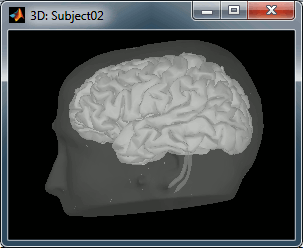
cortex_15000V: Low-resolution cortex surface, downsampled using the reducepatch function from Matlab (it keeps a meaningful subset of vertices from the original surface). This surface appears in green, it means that is going to be used as the default by the processes that require a cortex surface.
white_170000V: High-resolution white matter surface from BrainVISA (with MarsAtlas)
white_15000V: Low-resolution white matter, processed with reducepatch (with MarsAtlas)
A figure is automatically shown at the end of the process, in order to check visually that the low-resolution cortex and head surfaces were properly imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.
Handling errors
How to check the quality of the result
It's hard to estimate what would be a good cortical reconstruction. What you are trying to spot at this level is mostly the obvious errors, like when the early stages of the brain extraction didn't perform well, just with a visual inspection. Play with the Smooth slider in the Surface tab. If it looks like a brain (two separate hemispheres) in both smooth and original views, it is probably ok.
Display the cortex surface on top of the MRI slices, to make sure that they are well aligned, that the surface follows well the folds, and that left and right were not flipped: right-click on the low-resolution cortex > MRI registration > Check MRI/surface registration...
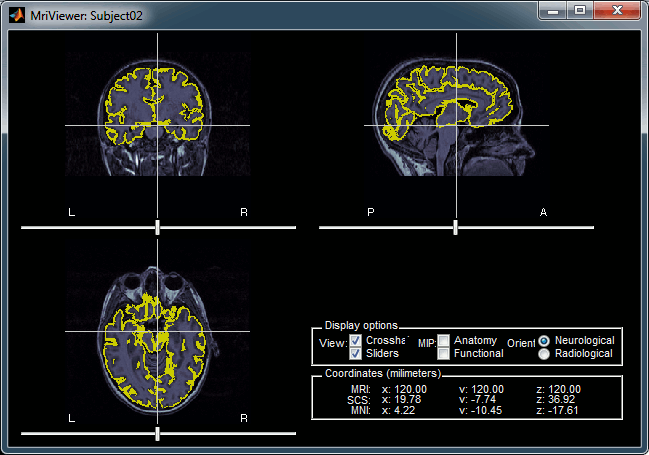
The cortex looks bad
It is critical to get a good cortex surface for source estimation. If the final cortex surface looks bad, it means that something didn't work well somewhere along the BrainVISA pipeline. The options are:
Ask help from the developer through the BrainVISA user forum
Use FreeSurfer instead
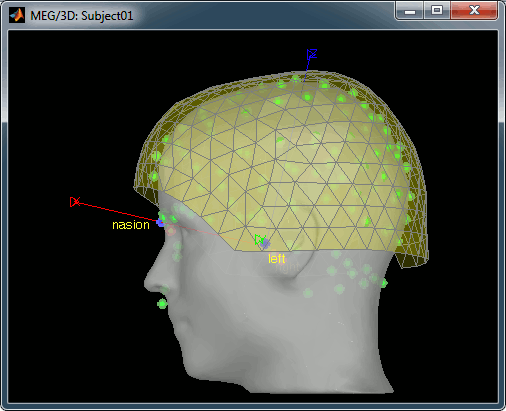
The head surface looks bad
It is not mandatory to have a perfect head surface to use any of the Brainstorm features: you don't necessarily have to recognize the face (for the anonymity of the figures, it can be even better if you don't).
The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can automatically align the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, these points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures.
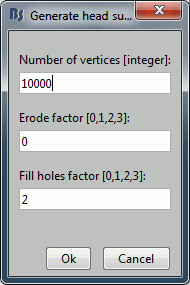
If the default head surface looks bad, you can try generating another one: right-click on the subject folder > Generate head surface. The options are:
Number of vertices: Number of points that are kept from the initial isosurface computed from the MRI. Increasing this number may increase the quality of the final surface.
Erode factor: Number of pixels to erode after the first binary threshold of the MRI. Increasing this number removes small components that are connected to the head.
Fill holes factor: Number of dimensions in which the holes should be identified and closed. Increasing this number removes more of the cavities of the head surface (0=no correction, 1=removes holes inside the surface, 3=closes all the features that make the surface non-convex)
Cortical parcellations
The process Cortical surface > Anatomy > Hip-Hop Cortical Parameterization in BrainVISA implements an automatic parcellation of the cortical surface in anatomical regions. The MarsAtlas model is described here:
https://meca-brain.org/software/marsatlas/
This atlas is available only on the white matter surface (ie. the grey-white interface, named white_*). It is imported in Brainstorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution white surface > File > View file contents. You can see the available structures "Atlas", the first one that has Name='User scouts', and the second one Name='MarsAtlas'.
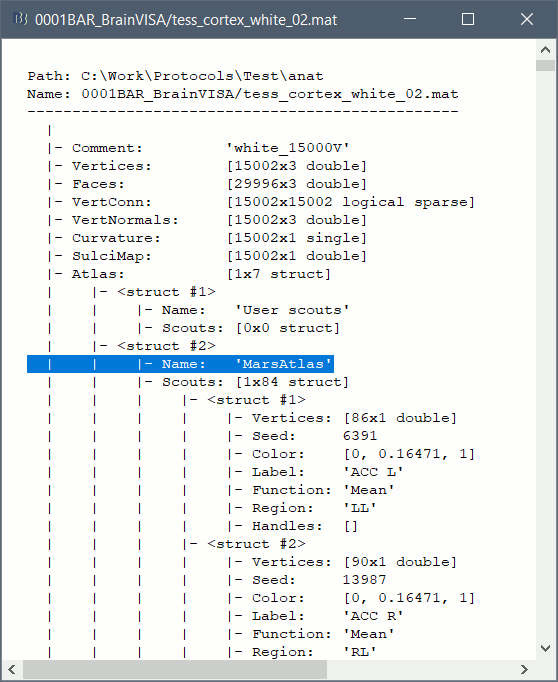
To access them from the interface: Double-click on the cortex and go to the Scout tab, and click on the drop-down list to select another Atlas (ie group of scouts):
Manual import of the anatomy
In case you need to import the MRI, surfaces and atlases separately instead of using the menu "Import anatomy folder", here is the sequence of operations to perform to get to the same result:
From the Anatomy side of the database explorer: create a subject.
Right-click on the subject folder > Import MRI > Select "nobias_subjectname.nii.gz"
- Set the 3 fiducial points NAS/LPA/RPA (or compute MNI normalization), save
Right-click on the subject folder > Import surfaces > Select simultaneously: head.gii, Lhemi.gii, Rhemi.gii, Lwhite.gii, Rwhite.gii
Double-click on Lwhite to display it. In the scout tab: Atlas > Load atlas > select the file format "GIfTI texture" and the file default_analysis/segmentation/mesh/surface_analysis/..._Lwhite_parcels_marsAtlas.gii. Close the figure.
- Repeat with surface Rwhite.
Select Lhemi, Rhemi, Lwhite, Rwhite, right-click > Less vertices > 7500 vertices > Select the first option "Matlab reducepatch"
Select Lhemi, Rhemi, right-click > Merge surfaces: Generates a surface cortex_70000V
Select Lwhite, Rwhite, right-click > Merge surfaces: Generates a surface white_70000V
Select Lhemi_7500V, Rhemi_7500V, right-click > Merge surfaces: Generates a surface cortex_15000V
Select Lwhite_7500V, Rwhite_7500V, right-click > Merge surfaces: Generates a surface white_15000V
- Delete all the separate hemispheres: *hemi*, *white*
- Double-click on cortex_15000V to set it as the default cortex