T1-MRI Segmentation with BrainVISA
It is not the purpose of Brainstorm tutorials to teach you how to use BrainVISA. But many Brainstorm users are lost when it gets to the segmentation of the MRI. So here is a short introduction to the BrainVISA T1 MRI processing pipeline. To extract head and cortex meshes from a T1 MRI, you can also try to use: BrainSuite or ?FreeSurfer.
We are going to illustrate the use of BrainVISA with the MRI from the CTF tutorials. You should already have those files on your computer, if you followed the basic tutorials. If it is not the case, go to the Download page, and get the file bst_sample_ctf.zip.
Installation
Download the latest version of BrainVISA from http://brainvisa.info
Install it on your computer by following the instructions on the download page.
- Start BrainVISA
You have to create a database for storing your files: this will be either asked at startup, or you'll have to select the menu !BrainVISA menu > Preferences:
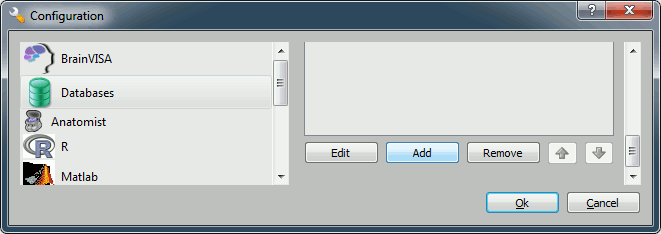
Click on Add, and create a directory with is not in any Brainstorm directory. For example:
Windows: My Documents\brainvisa_db\
Linux: /home/username/brainvisa_db/
MacOS: Documents/brainvisa_db/
- Click on Ok.
- Your are ready to import and process your MRI.
Import MRI
To process a MRI volume with BrainVISA, you have first to import it in the database.
Select the process: T1-MR1 > Import > Import T1 MRI, and click on Open button.
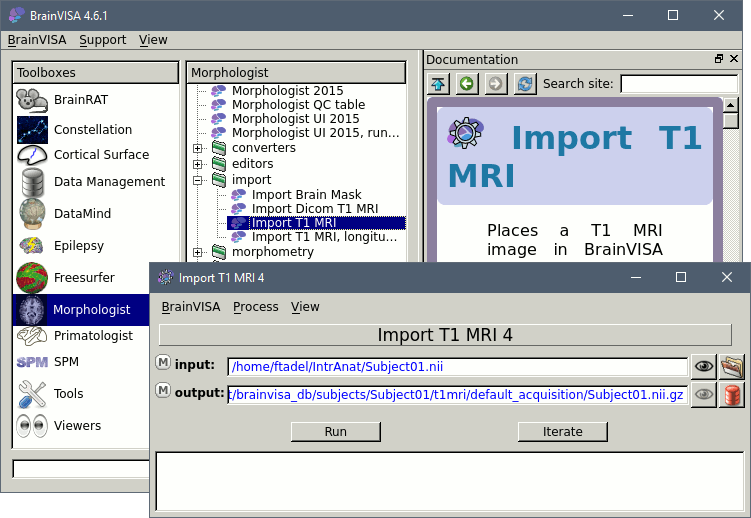
The following figure will appear. On the input line, click on the last button to select the file bst_tutorial_ctf/Anatomy/01.nii
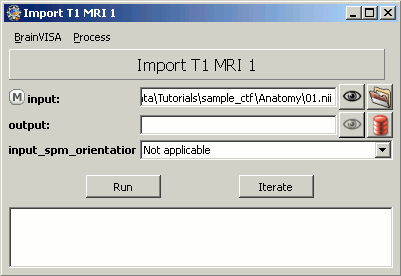
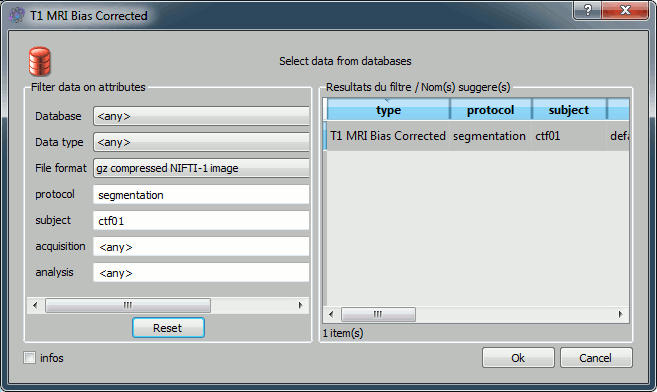
On the output line: click on the last button (red thing), which means: select an output file from the database. In the file selection window, you have to type the name of a new protocol, because there are no existing protocol yet. Type "Segmentation" in the protocol text box. You should see the default filenames appearing in the list on the right of the window. Click on Ok.
- Click on Run, wait for the process to end. Then you can close this window.The MRI is imported in the database.
Configure T1 pipeline
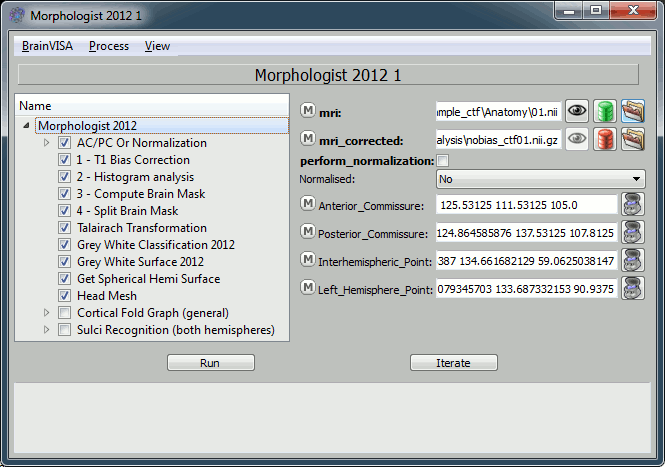
Back to BrainVISA window, now select process T1 MRI > Segmentation Pipeline > T1 Pipeline.
Click on open.
How to fill all those fields:
- Mri: click on the green button, and select from the database the MRI you've just imported
- Anterior commissure:
- Click on the "Anatomist" button (at the end of the line). This will start anatomist.
- In Anatomist window, look for a view that gives you an axial view
Then place the cross on the anterior commissure (see page: CoordinateSystems)
- Go back to the "T1 Pipeline" window again, and click again on "Anatomist" button for the anterior commissure. This should write the current position of the cursor in the "Anterior commissure" field.
- Repeat for all the other points: Posterior commissure, Interhemispheric point, Left hemisphere point
- You should pick the "left hemisphere" point in what appears to be the left hemisphere in the anatomist window, ie. the left side of the window.
- Uncheck the "Cortical Fold Graph" step, we don't need that.
Go to the "AC/PC Or Normalization" step, and select true for the option allow_flip_initial_MRI.
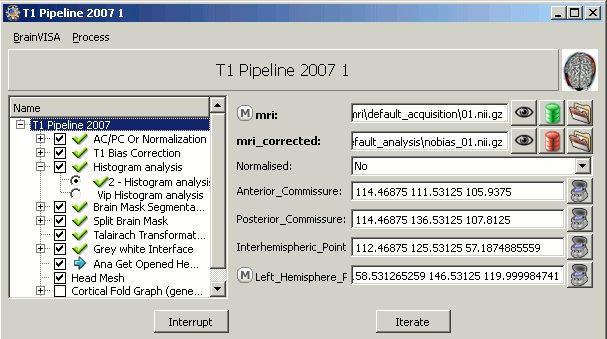
Click on Run and pray hard.
What if it crashes ?
Your first try might be a failure. Don't give up, it might be easy to fix.
If the process that crashed has two option (like all the steps from "T1 Bias correction" to "Split brain mask"):
- Select the other option (Example: "VIP bias correction" instead of "1 - T1 Bias correction")
- Uncheck all the processes that were successful (indicated with a green arrow), so that the Pipeline do not have to start from scratch again.
- Click on Run again.
If it still crashes:
- Try to change other options (like the selections in the previous steps)
- Post your anger about this absolute waste of time on BrainVISA forum
- Try another software.
And what if it worked ?
If you see at some point green arrows in front of all the processes: you are done.
You now need to import the files BrainVISA produced in BrainStorm. Here are the paths on my installation of BrainVISA (can be slightly different on your computer...):
MRI: BrainVisa_db\Segmentation\4D\t1mri\default_acquisition\default_analysis\nobias_4D.nii.gz
- I recommend to use the nobias volume, as you are sure that it will be correctly oriented in Brainstorm.
- If Brainstorm still doesn't have a .nii.gz format in Import MRI window, you'll have to gunzip this file first.
Scalp: BrainVisa_db\Segmentation\4D\t1mri\default_acquisition\default_analysis\segmentation\mesh\4D_head.mesh
Left hemisphere: BrainVisa_db\Segmentation\4D\t1mri\default_acquisition\default_analysis\segmentation\mesh\4D_Lhemi.mesh
Right hemisphere: BrainVisa_db\Segmentation\4D\t1mri\default_acquisition\default_analysis\segmentation\mesh\4D_Rhemi.mesh
- Remind to always import ALL the surface at the SAME time in Brainstorm, if not they won't be correctly registered with the MRI.
Never forget to check the final surfaces in Brainstorm, they still might be completely wrong. Example of a first try with BrainVISA, after import + merge.
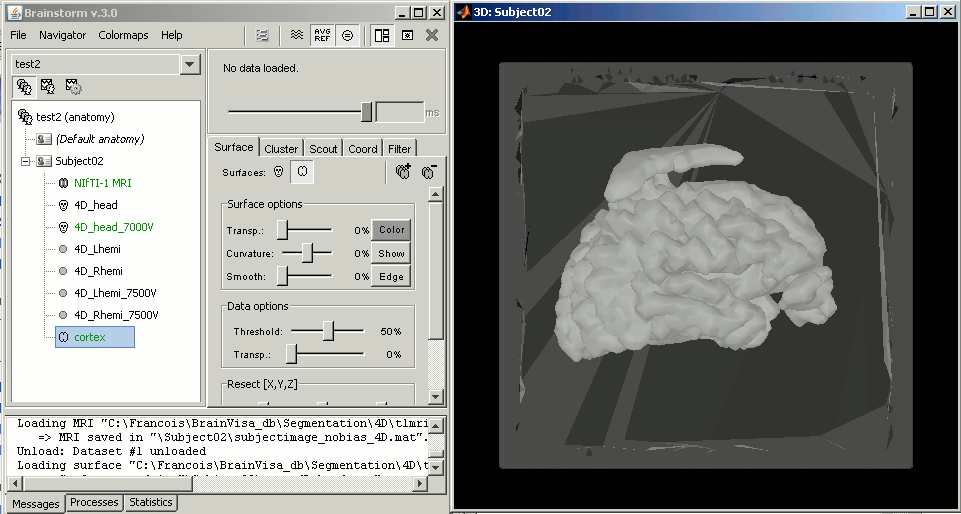
If this happen to you too, you'll have to start again the whole segmentation process, either selecting other options in BrainVISA, or using another software solution.
Have fun...