|
Size: 27412
Comment:
|
Size: 32357
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 107: | Line 107: |
== Electrode labeling and contact localization == === Generate isoSurface === This creates a thresholded mesh from the CT by separating the contacts out from rest of the CT. This aids the user towards localization of the electrodes and its contacts more accurately. * The first thing required is to define a good threshold (which we call '''isoValue''') that will separate out the contacts from the rest of the CT. Just open the CT file (by double clicking on it), click on the '''MIP: Functional''' in the MRI Viewer, use the '''Data Options > Amplitude''' slider in the '''Surface''' tab to browse through the threshold, and once you are satisfied with getting a good separation of the contacts, hover on the slider to get the desired value and keep a note of it (to the nearest integer). In this case the value is '''2276'''. <<BR>><<BR>> {{attachment:8_choose_isovalue.png||width="500"}} * Right click on '''post_CT_spm_reslice_masked_spm> CT segmentation > Generate SEEG/ECoG isosurface'''.<<BR>><<BR>> {{attachment:9_isosurface_menu.png||width="350"}} * This will bring the '''Generate isosurface''' window. This window shows 4 values: the '''Background level''', '''White level''', '''Max Intensity''' and the suggested '''Set isoValue''', all of them given in the [[https://en.wikipedia.org/wiki/Hounsfield_scale|Hounsfield Unit (HU) scale]]. The first 3 values are calculated automatically from the histogram of the CT and are displayed for reference. The editable '''isoValue''' field shows an estimated best guess based on mean of White Level and Max Intensity. Since we got a better value in step-1 above, set the value to '''2276''' and press '''OK'''.<<BR>><<BR>> {{attachment:10_isosurface_popup.png||width="350"}} * An isosurface is generated showing the contact as blobs overlayed on the 3D MRI slices. The '''Thresh''' slider under '''Surface options''' can be further used to fine tune and regenerate mesh with different isoValues.<<BR>><<BR>> {{attachment:11_isosurface_display.png||width="650"}} === Start implantation === * Right click on '''Subject01''', and choose '''SEEG/ECOG implantation'''. * The '''SEEG/ECOG implantation''' menu pops to choose which modalities you want to use for doing your implantation with. Choose '''MRI+CT+IsoSurf'''. This takes you to the functional tab. '''Subject01 > Implantation > SEEG/ECOG (0)''' channel gets created. An MRI Viewer loads up with the CT overlayed on the MRI. A 3D figure loads up with isoSurface and 3D MRI. '''Panel iEEG''' loads up in the panel viewer. Move around the 3D slices to get a clear view of the contacts so that they can be clicked on. See the section [[https://neuroimage.usc.edu/brainstorm/Tutorials/SeizureFingerprinting#SEEG.2FECOG_Implantation_Menu|SEEG/ECOG Implantation Menu]] for more details on this option.<<BR>><<BR>> {{attachment:12_implant_menu.png}} <<BR>><<BR>> {{attachment:13_implant_setup.png||width="650"}} {{{#!wiki note Go to section [[https://neuroimage.usc.edu/brainstorm/Tutorials/SeizureFingerprinting#Panel_iEEG|Panel iEEG]] to know more about the panel and its features. }}} === Create electrodes and plot contacts manually === * After performing steps above, we will have the Panel iEEG along with the MRI Viewer and 3D figure open. * Before we start, we need to note the names of the channels (which are also the contacts) that are there in the '''EDF channels''' above. The reason we do this is once we do the implantation and define the contact locations, we will import these positions to the '''EDF channels''' and Brainstorm does this by matching the channels by name. * Right click on '''EDF channels > Edit channel file''' to open the channel editor that lists all the channel in details. <<BR>><<BR>> {{attachment:16_edit_channel.png|width="650"}} * To summarize having this info along with the implantation scheme will help in doing the further steps. == Panel iEEG == |
* Select a scaling factor: '''1'''<<BR>>The positions in this text file are in millimeters, the expected unit for this file format is also millimeters, there is no adjustment to make here.<<BR>><<BR>> {{attachment:18_import_pos_factor.png||width="500"}} * For the reference MRI, since the '''Subject01_electrodes.tsv''' file consists of contact positions which were localized on the '''post_CT''' after it was coregistered to the '''pre_T1''', resliced and skull-stripped within Brainstorm, so selecting either of them as the reference MRI would be fine as they are both in the same space. Since we localized using the contacts from CT , select '''post_CT_spm_reslice_masked_spm'''.<<BR>><<BR>> {{attachment:19_import_pos_ref.png||width="250"}} * At the end, you get a report indicating how many channels from the SEEG recordings were attributed a new 3D position. The channels are matched by name: the position file you import must include the labels of the channels and they must be named exactly in the the same way as in your recordings.<<BR>><<BR>> {{attachment:20_import_pos_report.png||width="350"}} * You should always validate that the type of all the channels has been detected correctly. Check also that the names are correct and using the same convention for all the contact of a given electrode. These are entered manually, and typing errors are frequent. Right-click on a channel file > '''Edit channel file'''.<<BR>><<BR>> {{attachment:16_edit_channel.png||width="500"}} * To edit one channel, double-click on the text to modify. To edit multiple channels, select them all and right-click > Set channel group/type. The column Group corresponds to the name of the depth electrode. It is detected based on the channel type and name. If you include as SEEG a channel that detected as something else or rename a channel, you would need to manually update the channel name. * For '''EDF Channels''' under '''Baseline''' and '''ictal_repetitive_spike''', there were 3 channels not found. This is because of wrong channel information in the EDF files. In the '''Channel Editor''' you can see that the locations were not imported for these 3 channels. Edit the respective channel files (one at a time) and do the following: * Double-click the Name '''SEEGMPS1''' on channel '''95''' and rename it to '''MPS1''' * Double-click the Name '''SEEG_NO_LOCMPS1_02''' on channel '''109''' and rename it to '''MPS15''' * Double-click the Name '''SEEG_NO_LOCMPS1''' on channel '''110''' and rename it to '''MPS16''' * Select the channels '''95, 109, 110''' (Ctrl+click the individual channels), right click and set channel type as '''SEEG''' and channel group as '''MPS'''. * Close the channel editor and click '''Yes''' to save the modifications. * Select the two channel files and then import the contact positions from the '''Subject01_electrodes.tsv''' file again as we did above and now all missing locations will be imported correctly.<<BR>><<BR>> {{attachment:21_import_pos_report1_correct.png||width="350"}} * For '''EDF Channels''' under '''LVFA_and_wave''' and '''interictal_spike''', there were 15 channels not found. Out of the 15, 12 were EEG data (channels '''245-255, 262''') so they were correctly set to '''EEG_NO_LOC''' to ignore them when processing the SEEG data. The remaining 3 was because of wrong channel information in the EDF files. In the '''Channel Editor''' you can see that the locations were not imported for these 3 channels. Edit the respective channel files (one at a time) and do the same the we did in the above case for channels '''95, 109, 110'''. Save the change and re-import the contact positions to update the missing locations.<<BR>><<BR>> {{attachment:22_import_pos_report2_correct.png||width="350"}} * If you don't have the positions for the SEEG contacts, or if they don't look correctly aligned, you can directly place them in the MRI viewer. See the section [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Edit_the_contacts_positions|Edit the contacts positions]]. == Display the depth electrodes == === 3D figures === * Right-click on the channel file > '''Display sensors''' and explore all the available options.<<BR>><<BR>>{{attachment:23_display_menu.png||width="500"}} * You can render the SEEG depth electrodes in 3D together with the subject anatomy: surfaces, pre- or post-implantation volumes. You can add more anatomy elements to the figure with the button '''Add a surface''' ({{attachment:iconSurfaceAdd.gif}}) at the top-right of the '''Surface''' tab. For more help: [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#MRI_in_3D|Display the anatomy]].<<BR>><<BR>>{{attachment:24_display_3d.png||width="650"}} * Click on a contact to select it, right-click on it to get its name.<<BR>><<BR>>{{attachment:25_display_selectchan.png||width="400"}} === MRI Viewer === === Panel iEEG === |
| Line 165: | Line 165: |
| <<TAG(Advanced)>> == Electrode labeling and contact localization (WIP) == === Generate isoSurface === This creates a thresholded mesh from the CT by separating the contacts out from rest of the CT. This aids the user towards localization of the electrodes and its contacts more accurately. * The first thing required is to define a good threshold (which we call '''isoValue''') that will separate out the contacts from the rest of the CT. Just open the CT file (by double clicking on it), click on the '''MIP: Functional''' in the MRI Viewer, use the '''Data Options > Amplitude''' slider in the '''Surface''' tab to browse through the threshold, and once you are satisfied with getting a good separation of the contacts, hover on the slider to get the desired value and keep a note of it (to the nearest integer). In this case the value is '''2276'''. <<BR>><<BR>> {{attachment:8_choose_isovalue.png||width="500"}} * Right click on '''post_CT_spm_reslice_masked_spm> CT segmentation > Generate SEEG/ECoG isosurface'''.<<BR>><<BR>> {{attachment:9_isosurface_menu.png||width="350"}} * This will bring the '''Generate isosurface''' window. This window shows 4 values: the '''Background level''', '''White level''', '''Max Intensity''' and the suggested '''Set isoValue''', all of them given in the [[https://en.wikipedia.org/wiki/Hounsfield_scale|Hounsfield Unit (HU) scale]]. The first 3 values are calculated automatically from the histogram of the CT and are displayed for reference. The editable '''isoValue''' field shows an estimated best guess based on mean of White Level and Max Intensity. Since we got a better value in step-1 above, set the value to '''2276''' and press '''OK'''.<<BR>><<BR>> {{attachment:10_isosurface_popup.png||width="350"}} * An isosurface is generated showing the contact as blobs overlayed on the 3D MRI slices. The '''Thresh''' slider under '''Surface options''' can be further used to fine tune and regenerate mesh with different isoValues.<<BR>><<BR>> {{attachment:11_isosurface_display.png||width="650"}} === Start implantation === * Right click on '''Subject01''', and choose '''SEEG/ECOG implantation'''. * The '''SEEG/ECOG implantation''' menu pops to choose which modalities you want to use for doing your implantation with. Choose '''MRI+CT+IsoSurf'''. This takes you to the functional tab. '''Subject01 > Implantation > SEEG/ECOG (0)''' channel gets created. An MRI Viewer loads up with the CT overlayed on the MRI. A 3D figure loads up with isoSurface and 3D MRI. '''Panel iEEG''' loads up in the panel viewer. Move around the 3D slices to get a clear view of the contacts so that they can be clicked on. See the section [[https://neuroimage.usc.edu/brainstorm/Tutorials/SeizureFingerprinting#SEEG.2FECOG_Implantation_Menu|SEEG/ECOG Implantation Menu]] for more details on this option.<<BR>><<BR>> {{attachment:12_implant_menu.png}} <<BR>><<BR>> {{attachment:13_implant_setup.png||width="650"}} {{{#!wiki note Go to section [[https://neuroimage.usc.edu/brainstorm/Tutorials/SeizureFingerprinting#Panel_iEEG|Panel iEEG]] to know more about the panel and its features. }}} === Create electrodes and plot contacts manually === * After performing steps above, we will have the Panel iEEG along with the MRI Viewer and 3D figure open. * Before we start, we need to note the names of the channels (which are also the contacts) that are there in the '''EDF channels''' above. The reason we do this is once we do the implantation and define the contact locations, we will import these positions to the '''EDF channels''' and Brainstorm does this by matching the channels by name. * Right click on '''EDF channels > Edit channel file''' to open the channel editor that lists all the channel in details. <<BR>><<BR>> {{attachment:16_edit_channel.png|width="500"}} * To summarize having this info along with the implantation scheme will help in doing the further steps. <<TAG(Advanced)>> |
|
| Line 175: | Line 199: |
SEEG Time-Frequency Fingerprint Analysis for Epileptogenic Zone Localization (under construction)
Authors: xxx.
This tutorial introduces some concepts that are specific to the management of intracranial, SEEG recordings in the Brainstorm environment, and explains how to compute maps of epileptogenicity from ictal and interictal recordings. It is based on a clinical case from the McGovern Medical School, University of Texas Health Science Center at Houston, Texas, USA.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
NOT FOR CLINICAL USE:
The performance characteristics of the methods and software implementation presented in this tutorial have not been certified as medical devices and should be used for research purposes only.
Contents
Dataset description
License
This tutorial dataset (EEG, MRI and CT data) remains property of the McGovern Medical School, University of Texas Health Science Center at Houston, Texas, USA. Its use and transfer outside the Brainstorm tutorials, e.g. for research purposes, is prohibited without written consent. For questions, please contact Yash Shashank Vakilna, MS ( Yash.Shashank.Vakilna@uth.tmc.edu ).
Clinical description
The dataset was recorded at the Epilepsy Monitoring Unit at UTHealth Houston. It includes recordings for a patient who was a 25-year-old right-handed woman with drug-resistant epilepsy since age six and a prior right parietal opercular corticectomy at 15 presented with weekly focal aware seizures featuring a left-hand tingling aura and focal impaired awareness seizures with staring and pouting. In the Epilepsy Monitoring Unit (EMU) she had intermittent right parietal slowing and ten habitual seizures arising from C4-P4, and MRI revealed bilateral perisylvian polymicrogyria (PMG), pachygyria, right posterior temporal periventricular nodular heterotopia, and post-surgical changes. MEG localized discharges to the right superior parietal region adjacent to her previous resection, and SEEG implantation mapped two distinct onset patterns: low-voltage fast activity in right superior parietal PMG during focal aware seizures and repetitive spiking in posterior insular PMG during impaired awareness seizures. After multidisciplinary review, she underwent uncomplicated MR-guided laser interstitial thermal therapy targeting the right superior parietal and posterior insular PMG and remained seizure-free at one-year follow-up.
SEEG recordings
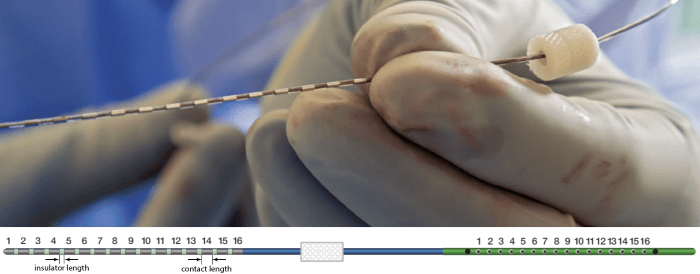
The depth electrodes used in this example dataset are PMT SEEG Depth Electrodes, with the following specifications:
- Diameter: 0.8 mm
- Contact length: 2 mm
- Insulator length: 1.5 mm
- Distance between the center of two contacts: 3.5 mm
- Between 8 and 16 contacts on each electrode
Files
tutorial_seizure_fingerprinting/
anatomy/: Anatomy data
pre_T1.nii.gz: Raw pre-implantation T1 MRI (in NIfTI-1 format)
pre_T2_FLAIR.nii.gz: Raw pre-implantation T2 FLAIR MRI (in NIfTI-1 format)
post_CT.nii.gz: Raw post-implantation CT scan (in NIfTI-1 format)
post_CT_coreg.nii.gz: Post-implantation CT scan (in NIfTI-1 format) coregistered, resliced and skull-stripped using SPM
pre_DWI/pre_DWI.nii.gz: Raw diffusion weighted MRI (in NIfTI-1 format)
pre_DWI/pre_DWI.bval: Contains a single number per volume that indicates how large of a diffusion gradient was applied to the data
pre_DWI/pre_DWI.bvec: Contains a triplet of numbers per volume that shows in what directions the gradients were applied
recordings/: SEEG recordings
Baseline.edf: Raw SEEG recordings (in EDF format) for baseline
ictal_repetitive_spike.edf: Raw SEEG recordings (in EDF format) for seizure with Ictal repetitive spiking
interictal_spike.edf: Raw SEEG recordings (in EDF format) for interictal spike
LVFA_and_wave.edf: Raw SEEG recordings (in EDF format) for seizure onset with Low-voltage-fast-activity
Subject01_electrodes.tsv: Position of contacts localized using Brainstorm
References
All details for this study can be found here: https://zenodo.org/records/14807262
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
BrainSuite (optional): Make sure you have the latest version of BrainSuite installed. Follow steps as per the BrainSuite for Brainstorm tutorial. This is required to perform skull stripping when importing CT below in order to remove extracranial regions from the CT.
SPM: If you are running Brainstorm from the MATLAB environment, you need to have the SPM12 toolbox installed on your computer, as a Brainstorm plugin or a custom installation.
With the stand-alone compiled version of Brainstorm: all the needed SPM scripts have been compiled and included in the executable. This is required for two purposes:- Coregistration between pre-implantation MRI and post-implantation CT volumes.
Use mask from SPM based tissue segmentation to remove extracranial regions from the CT.
ct2mrireg: To have an alternate way of doing the coregistration between pre-implantation MRI and post-implantation CT volumes, you need to have the ct2mrireg plugin installed on your computer, as a Brainstorm plugin. It can be found under the menu Plugins > Anatomy > ct2mrireg. If using Brainstorm with MATLAB, please install the Image Processing Toolbox.
Download the dataset:
Go to the Download page of this website, and download the file: tutorial_seizure_fingerprinting.zip.
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder).
Brainstorm:
- Start Brainstorm
Select the menu File > Create new protocol. Name it "TutorialSeizureFingerprinting" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
Import the anatomy
Pre-implantation MRI
Right-click on the TutorialSeizureFingerprinting folder > New subject > Subject01.
Keep the default options you defined for the protocol.Switch to the Anatomy view of the protocol.
Right-click on the subject node > Import MRI.
Set the file format: MRI: NIfTI-1 (*.nii;*.nii.gz).
Select: tutorial_seizure_fingerprinting/anatomy/pre_T1.nii.gzThe MRI viewer opens automatically. Click on "Click here to compute MNI normalization", option "maff8". This method is embedded in Brainstorm and does not require the installation of SPM12. However, it requires the automatic download of the file SPM12 Tissue Probability Maps. If you do not have access to internet, see the instructions on the Installation page. It is based on an affine co-registration with the MNI ICBM152 template from the SPM software, described in the following article: Ashburner J, Friston KJ, Unified segmentation, NeuroImage 2005..
Click on Save to close the MRI viewer. New node named pre_T1 is created.

While it is not applicable to this data, but while importing some MRIs if there is a transformation available in the NIfTI header, then a window pops up asking Do you would want to apply the transformation to the MRI file ? Choosing Yes will orient the MRI based on this transformation and will reorient the MRI in Brainstorm's standard orientation, so you can see the coronal/sagittal/axial views correctly oriented. More details.
Post-implantation CT
The pre-implantation MRI above will be used as the anatomical reference for this subject. We will now import a second scan done after the SEEG implantation, on which we can see the SEEG contacts. In this dataset, the post-implantation volume is a CT scan (contacts hypersignal appear in white).
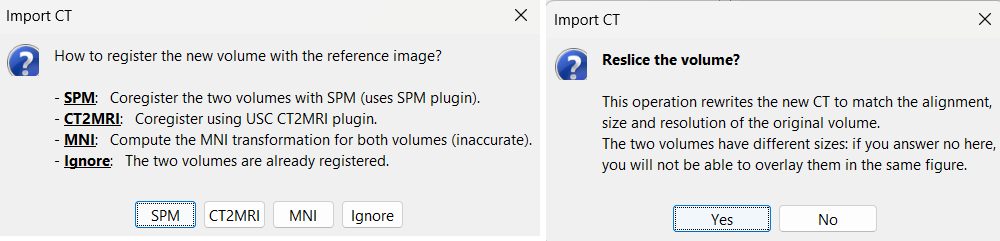
Right-click on the subject node > Import CT.
Select: tutorial_seizure_fingerprinting/anatomy/post_CT.nii.gzChoose SPM for coregistration. See the section Volume coregistration for more details on this option.
Choose Yes for reslicing so that the CT voxel dimensions match those of the MRI.
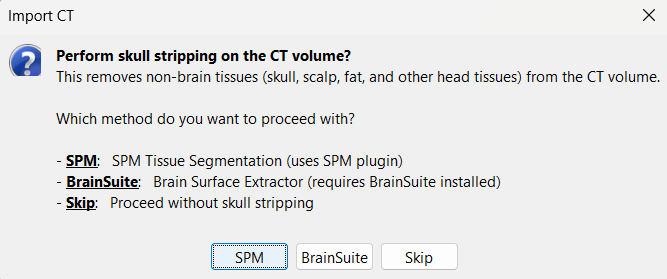
Choose SPM for skull stripping to remove any non-brain tissues as we need just the electrodes inside the brain. See the section Skull Stripping for more details on this option.
The MRI viewer opens automatically, showing the post-implantation CT volume as a colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section Data options of the Surface tab, adjust its colormap with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well.
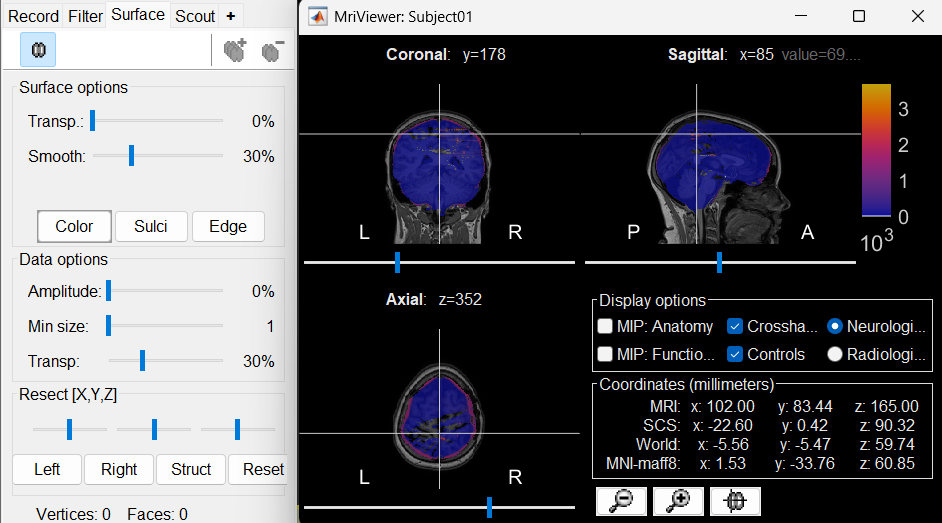
New node named post_CT_spm_reslice_masked_spm is created. The postfix _spm_reslice_masked_spm indicates the different processes it has gone through. To see the history of the processes for this file, right click on post_CT_spm_reslice_masked_spm > File > View file history.
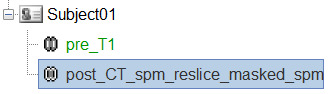
To just view the CT File, right click on post_CT_spm_reslice_masked_spm > Display > MRI Viewer.
Generate default surfaces using CAT12
We recommend generating cortical surfaces with CAT12, especially if you are interested in a realistic representation of the patient's cortical folding in 3D. Follow the CAT12 tutorial to generate the surfaces as under.
These surfaces will be used later, in the computation of the epileptogenicity maps. Read the section Importing realistic surfaces for information on how to use realistic surfaces from BrainVISA or FreeSurfer.
Access the recordings
Link the recordings
- Switch to the "functional data" view (2nd button, on top of the database explorer).
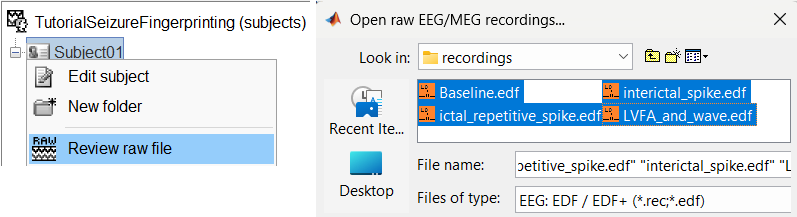
Right-click on the subject folder > Review raw file:
Select the file format: EEG: EDF/EDF+
Select all the recordings: tutorial_seizure_fingerprinting/recordings/*.edf
The new files Link to raw file let you access directly the contents of the original SEEG files. The menu Review raw file does not actually copy any data to the database. More details.
Import the contacts positions
In order to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the depth electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. To make this tutorial easier to reproduce and follow, we distribute the positions of the contacts saved in a text file. For instructions to manually localize the SEEG contacts using Brainstorm, read the advanced section Electrode labeling and contact localization (WIP).
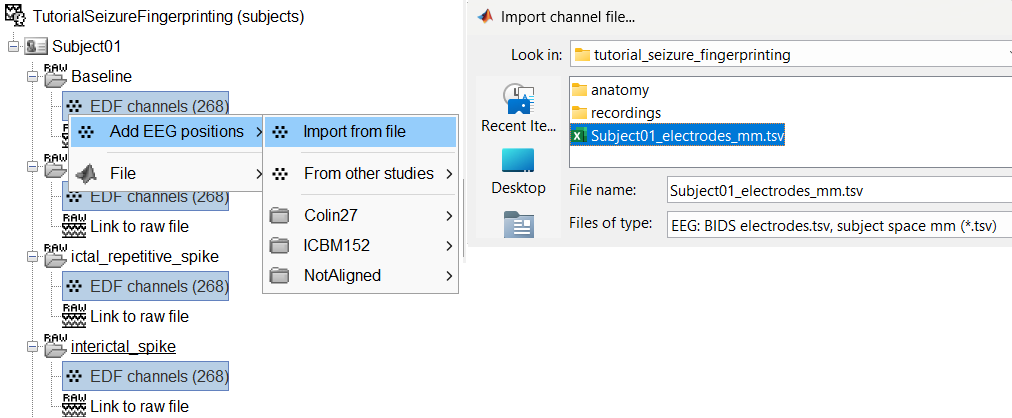
- The channel file "EDF channels" contain the name of the channels, but not their positions. We need to import or edit separately the positions of the SEEG contacts.
Click on the [+] next to the four folders, select all the channel files simultaneously.
Right-click one channel file > Add EEG positions > Import from file
Select format: EEG: BIDS electrodes.tsv, subject space mm (*.tsv)
Select file: tutorial_seizure_fingerprinting/Subject01_electrodes.tsv
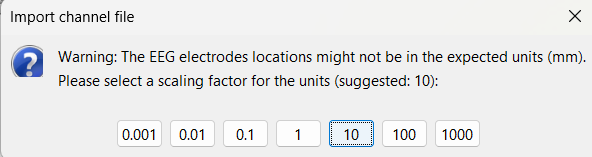
Select a scaling factor: 1
The positions in this text file are in millimeters, the expected unit for this file format is also millimeters, there is no adjustment to make here.
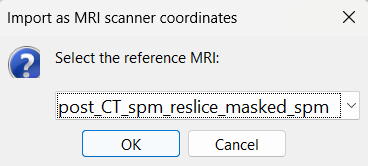
For the reference MRI, since the Subject01_electrodes.tsv file consists of contact positions which were localized on the post_CT after it was coregistered to the pre_T1, resliced and skull-stripped within Brainstorm, so selecting either of them as the reference MRI would be fine as they are both in the same space. Since we localized using the contacts from CT , select post_CT_spm_reslice_masked_spm.
At the end, you get a report indicating how many channels from the SEEG recordings were attributed a new 3D position. The channels are matched by name: the position file you import must include the labels of the channels and they must be named exactly in the the same way as in your recordings.
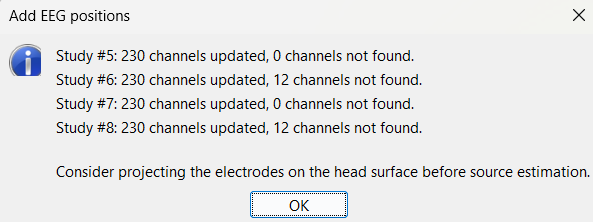
You should always validate that the type of all the channels has been detected correctly. Check also that the names are correct and using the same convention for all the contact of a given electrode. These are entered manually, and typing errors are frequent. Right-click on a channel file > Edit channel file.
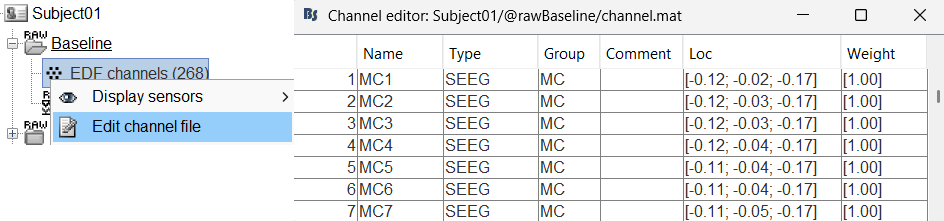
To edit one channel, double-click on the text to modify. To edit multiple channels, select them all and right-click > Set channel group/type. The column Group corresponds to the name of the depth electrode. It is detected based on the channel type and name. If you include as SEEG a channel that detected as something else or rename a channel, you would need to manually update the channel name.
For EDF Channels under Baseline and ictal_repetitive_spike, there were 3 channels not found. This is because of wrong channel information in the EDF files. In the Channel Editor you can see that the locations were not imported for these 3 channels. Edit the respective channel files (one at a time) and do the following:
Double-click the Name SEEGMPS1 on channel 95 and rename it to MPS1
Double-click the Name SEEG_NO_LOCMPS1_02 on channel 109 and rename it to MPS15
Double-click the Name SEEG_NO_LOCMPS1 on channel 110 and rename it to MPS16
Select the channels 95, 109, 110 (Ctrl+click the individual channels), right click and set channel type as SEEG and channel group as MPS.
Close the channel editor and click Yes to save the modifications.
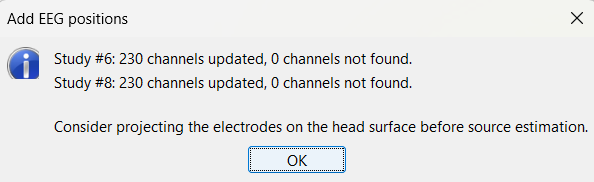
Select the two channel files and then import the contact positions from the Subject01_electrodes.tsv file again as we did above and now all missing locations will be imported correctly.
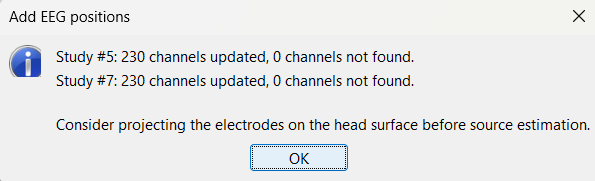
For EDF Channels under LVFA_and_wave and interictal_spike, there were 15 channels not found. Out of the 15, 12 were EEG data (channels 245-255, 262) so they were correctly set to EEG_NO_LOC to ignore them when processing the SEEG data. The remaining 3 was because of wrong channel information in the EDF files. In the Channel Editor you can see that the locations were not imported for these 3 channels. Edit the respective channel files (one at a time) and do the same the we did in the above case for channels 95, 109, 110. Save the change and re-import the contact positions to update the missing locations.
If you don't have the positions for the SEEG contacts, or if they don't look correctly aligned, you can directly place them in the MRI viewer. See the section Edit the contacts positions.
Display the depth electrodes
3D figures
Right-click on the channel file > Display sensors and explore all the available options.
You can render the SEEG depth electrodes in 3D together with the subject anatomy: surfaces, pre- or post-implantation volumes. You can add more anatomy elements to the figure with the button Add a surface (
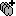 ) at the top-right of the Surface tab. For more help: Display the anatomy.
) at the top-right of the Surface tab. For more help: Display the anatomy.
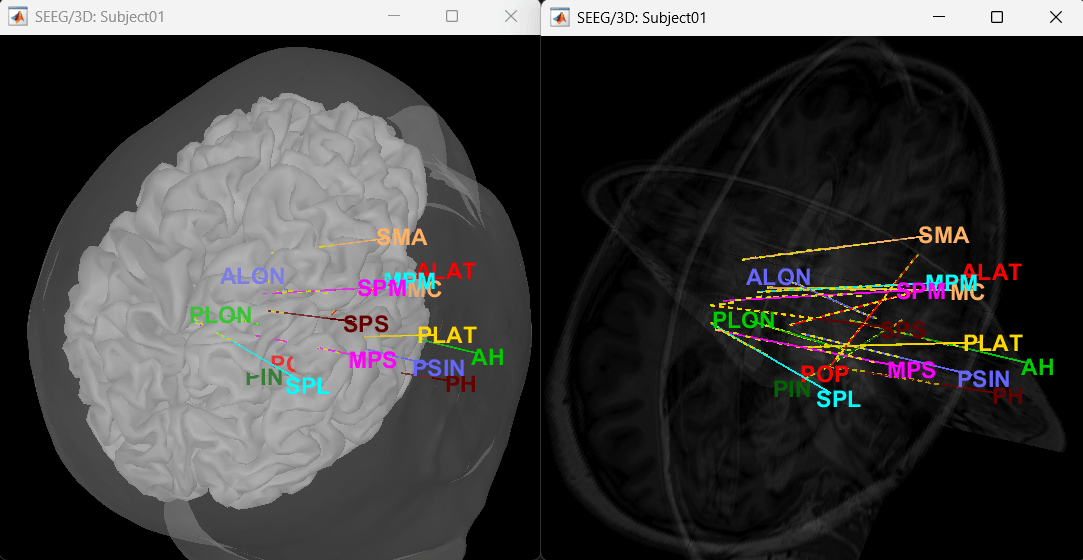
Click on a contact to select it, right-click on it to get its name.
MRI Viewer
Panel iEEG
When opening SEEG/ECOG recordings, the panel iEEG is added to the Brainstorm window. You can use it to edit the display properties of the depth electrodes. In this interface, "electrode" refers to entire depth electrode implanted in the head of the patient while "contact" refers to recording sites on the electrode. There are multiple contacts on an electrode, and one contact corresponds to one channel of data in the channel file and the recordings.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
SEEG depth electrodes are graphical objects, they are defined independently from the SEEG contacts available in the channel file and recordings. A contact/channel is associated to a depth electrode using the Group property of the channel, accessible with a right-click on the channel file > Edit channel file.
In this example, the depth electrodes available in the Panel iEEG have been manually set by using this Panel features. In the convention used here, the contact names must start with one or more letters (the name of the electrode) followed by a number representing the index of the contact on the electrode. Contact #1 is at the tip of the electrode, and is therefore the deeper contact of the electrode. If the convention used in your recordings is different, you may have to edit the electrodes properties in order to get them displayed correctly.
- Buttons in the toolbar:
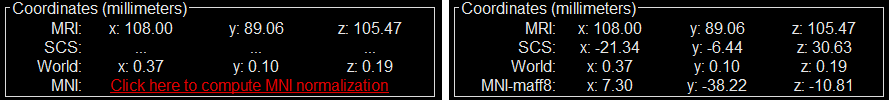
Coordinates (millimeters) radio buttons: Users can switch between the different coordinate systems (SCS, MRI, World, MNI) to change the coordinate values of the contacts.
 (Add new electrode): Adds an entry for a new depth electrode. The new electrode will not be displayed until you set its properties and position. This will not add or remove SEEG contacts or channels of data.
(Add new electrode): Adds an entry for a new depth electrode. The new electrode will not be displayed until you set its properties and position. This will not add or remove SEEG contacts or channels of data.  (Remove selected electrodes): Deletes a depth electrode from the list, but does not modify the list of SEEG contacts or channels of data.
(Remove selected electrodes): Deletes a depth electrode from the list, but does not modify the list of SEEG contacts or channels of data.  (Merge selected electrodes): Join two or more selected depth electrodes from the list. This will remove the selected electrodes after creating a new merged electrode with all their contacts combined in them. This will not modify the channels of the data (Shortcut: Ctrl+M).
(Merge selected electrodes): Join two or more selected depth electrodes from the list. This will remove the selected electrodes after creating a new merged electrode with all their contacts combined in them. This will not modify the channels of the data (Shortcut: Ctrl+M).  (Select surface point): Activates coordinate selection in 3D figure (Shortcut: Ctrl+P).
(Select surface point): Activates coordinate selection in 3D figure (Shortcut: Ctrl+P).  (Select surface centroid): Activates centroid selection of surface in 3D figure. This option activates for use (turns green) only when the
(Select surface centroid): Activates centroid selection of surface in 3D figure. This option activates for use (turns green) only when the  button is selected and the 3D figure has an IsoSurface present otherwise it appears grayed out (
button is selected and the 3D figure has an IsoSurface present otherwise it appears grayed out (  ). Use this to toggle between centroid and surface point selection in the IsoSurface.
). Use this to toggle between centroid and surface point selection in the IsoSurface.  (Set color for selected electrodes): Self explanatory.
(Set color for selected electrodes): Self explanatory.  (Show/hide selected electrodes): To hide an electrode in the 3D figures and MRI viewer, select it in the list then click on this button. Select all the electrodes with the standard shortcut Ctrl+A.
(Show/hide selected electrodes): To hide an electrode in the 3D figures and MRI viewer, select it in the list then click on this button. Select all the electrodes with the standard shortcut Ctrl+A. Display contacts as:
 (Depth electrodes) /
(Depth electrodes) /  (Spheres).
(Spheres). Contacts >
 Add SEEG contact: Adds a new contact to the selected SEEG electrode in the list. This will not modify the channels of the data (Shortcut: S).
Add SEEG contact: Adds a new contact to the selected SEEG electrode in the list. This will not modify the channels of the data (Shortcut: S). Contacts >
 Remove SEEG contacts: Deletes selected contacts from the list for an SEEG electrode, but does not modify the channels of the data (Shortcut: Delete).
Remove SEEG contacts: Deletes selected contacts from the list for an SEEG electrode, but does not modify the channels of the data (Shortcut: Delete). Contacts > Use default positions: For each of the selected electrodes, the current positions of the SEEG contacts are discarded and replaced with the default positions of the contacts on the electrode. The properties used for setting the position of the contacts are the contact spacing, the tip of the electrode and the entry point in the skull. Contact #i is placed along the electrode at (i-1)*contact_spacing millimeters from the tip of the electrode.
Contacts > Project on electrode: For each of the selected electrodes, the contacts are projected orthogonally on the electrode. This menu can be useful for aligning contacts that were marked one by one.
Contacts > Show/Hide line fit through contacts: Performs line fitting through the contacts in 3D figure.
Contacts > Save modifications: Save the current modifications to the channel file. Otherwise, the modifications are saved only when you close the figure (dialog box "Save modifications to channel file?")
Contacts > Export contacts positions: Save the 3D positions of the SEEG contacts in a text file, using one of the file formats supported by Brainstorm.
Electrode properties: Properties that have an impact on the position of the contacts.
- If you edit the properties, the modifications will apply to all the selected electrodes in the current channel file. Check what is selected before making changes.
Type: SEEG/ECOG
Model: List of electrode models. If you select an entry in this menu, it will copy the default properties for this model to the selected electrodes. If you are using electrodes that are not in this list, please post on the specification of your devices on the user forum and we will add them to this list.
Number of contacts: Number of recording sites on the electrode. By default, this is set for SEEG to the maximum index found in a group of contacts. Example: electrode s' is associated to channels s'1, s'2, s'3, s'10, s'11, s'12 => detected number of contacts is 12.
Contact spacing: Distance between the centers of two consecutive contacts in the electrode. In this example dataset, the default value corresponds to the average distance observed between pairs of adjacent contacts.
Set tip: Select one electrode in the list, then move the cursor of the MRI viewer to the tip of the selected electrode (center of the first contact), and finally click on [Set tip].
Set skull entry: Select one electrode in the list, then move the cursor of the MRI viewer to the point where the depth electrode enters the skull, and finally click on [Set skull entry]. This position does not correspond to any contact, it is used only to estimate the direction of the depth electrode. More details.
- Display options: Properties that only affect the way the electrodes are renderer graphically.
Contact length: Defines the length of the yellow cylinder that represents the contacts along the electrode axis, or the diameter of the sphere when the electrodes are not rendered.
Contact diameter: Diameter of the yellow cylinders representing the contacts. By default, this value is slightly larger than the electrode diameter so that it is rendered correctly. If you use the same value as the electrode diameter, the contacts might not be visible.
Electrode diameter: Diameter of the cylinder representing the SEEG depth electrode.
Electrode length: Length of the cylinder representing the SEEG depth electrode.
Electrode labeling and contact localization (WIP)
Generate isoSurface
This creates a thresholded mesh from the CT by separating the contacts out from rest of the CT. This aids the user towards localization of the electrodes and its contacts more accurately.
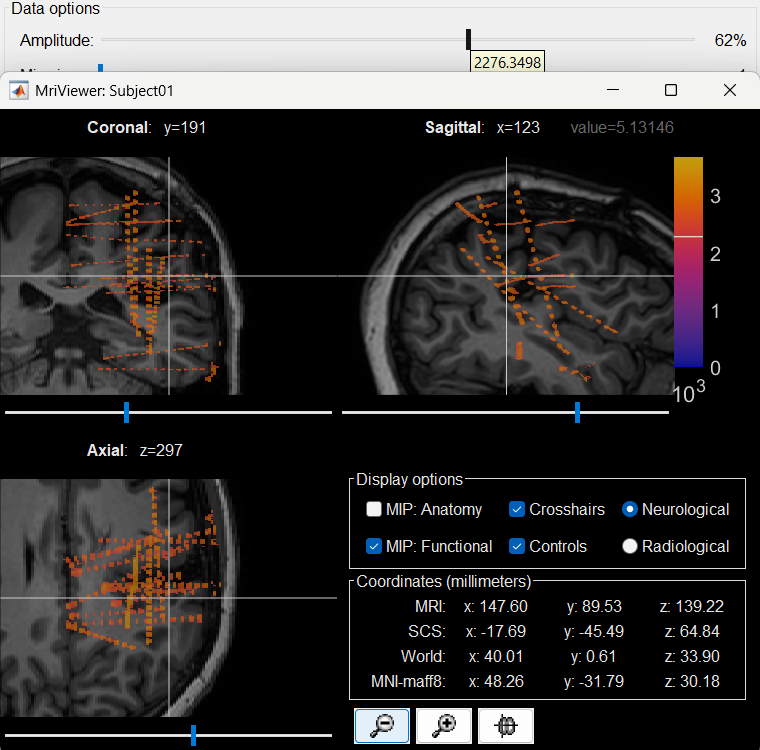
The first thing required is to define a good threshold (which we call isoValue) that will separate out the contacts from the rest of the CT. Just open the CT file (by double clicking on it), click on the MIP: Functional in the MRI Viewer, use the Data Options > Amplitude slider in the Surface tab to browse through the threshold, and once you are satisfied with getting a good separation of the contacts, hover on the slider to get the desired value and keep a note of it (to the nearest integer). In this case the value is 2276.
Right click on post_CT_spm_reslice_masked_spm> CT segmentation > Generate SEEG/ECoG isosurface.
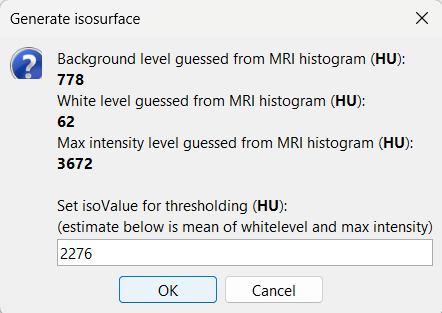
This will bring the Generate isosurface window. This window shows 4 values: the Background level, White level, Max Intensity and the suggested Set isoValue, all of them given in the Hounsfield Unit (HU) scale. The first 3 values are calculated automatically from the histogram of the CT and are displayed for reference. The editable isoValue field shows an estimated best guess based on mean of White Level and Max Intensity. Since we got a better value in step-1 above, set the value to 2276 and press OK.
An isosurface is generated showing the contact as blobs overlayed on the 3D MRI slices. The Thresh slider under Surface options can be further used to fine tune and regenerate mesh with different isoValues.

Start implantation
Right click on Subject01, and choose SEEG/ECOG implantation.
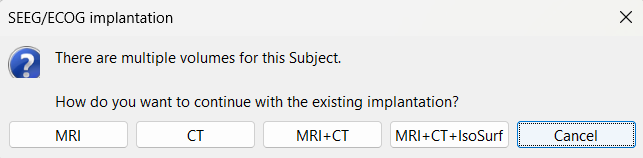
The SEEG/ECOG implantation menu pops to choose which modalities you want to use for doing your implantation with. Choose MRI+CT+IsoSurf. This takes you to the functional tab. Subject01 > Implantation > SEEG/ECOG (0) channel gets created. An MRI Viewer loads up with the CT overlayed on the MRI. A 3D figure loads up with isoSurface and 3D MRI. Panel iEEG loads up in the panel viewer. Move around the 3D slices to get a clear view of the contacts so that they can be clicked on. See the section SEEG/ECOG Implantation Menu for more details on this option.
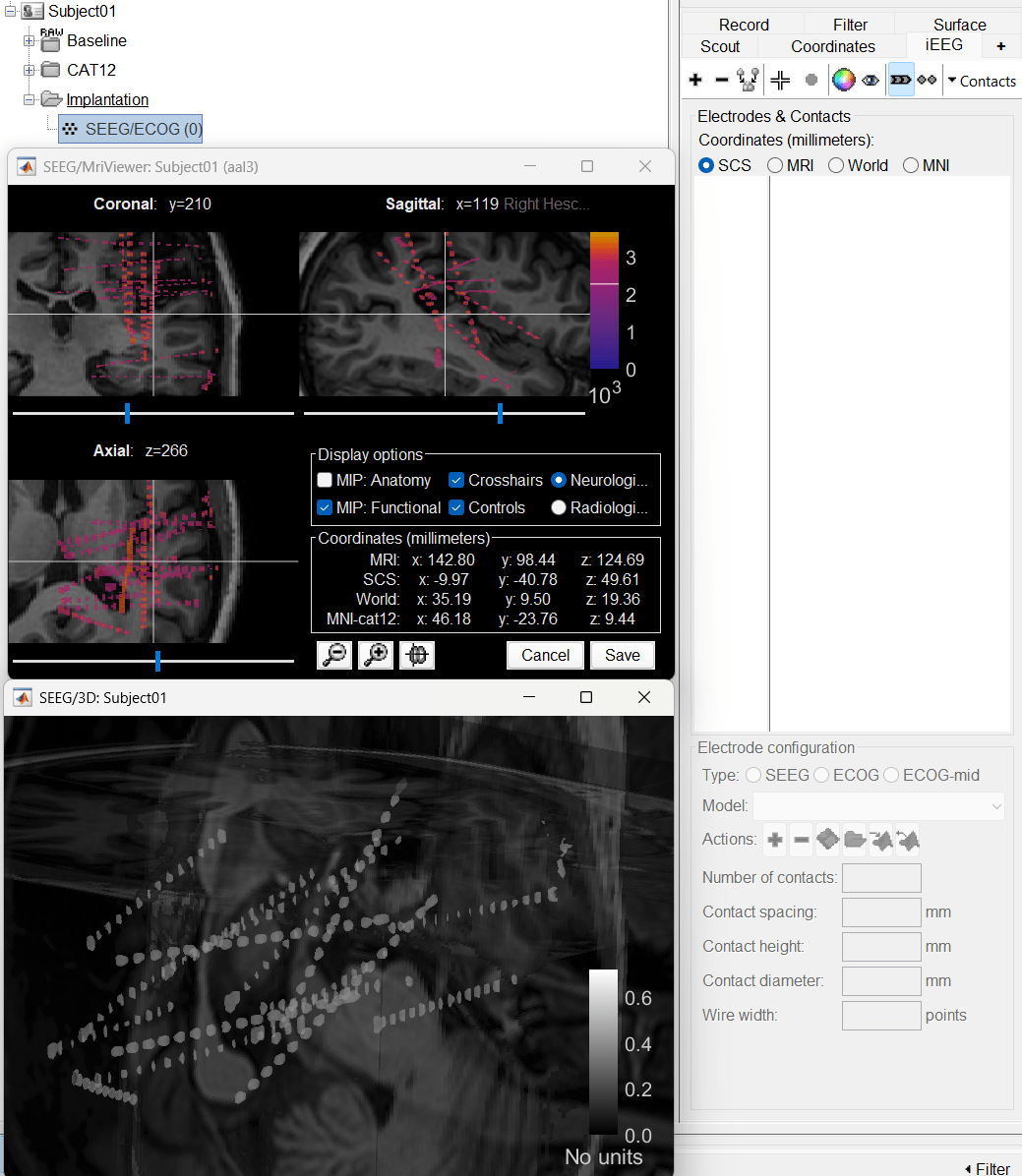
Go to section Panel iEEG to know more about the panel and its features.
Create electrodes and plot contacts manually
- After performing steps above, we will have the Panel iEEG along with the MRI Viewer and 3D figure open.
Before we start, we need to note the names of the channels (which are also the contacts) that are there in the EDF channels above. The reason we do this is once we do the implantation and define the contact locations, we will import these positions to the EDF channels and Brainstorm does this by matching the channels by name.
Right click on EDF channels > Edit channel file to open the channel editor that lists all the channel in details.

- To summarize having this info along with the implantation scheme will help in doing the further steps.
SEEG/ECOG Implantation Menu
While starting manual implantation on an volume by right click on subject node > SEEG/ECOG implantation, the menu below pops up allowing the user to choose which modality or modalities to proceed with.

MRI: MRI viewer loads up with MRI volume only.
CT: MRI viewer loads up with CT volume only.
MRI+CT: MRI viewer loads up with the CT overlayed on the MRI.
MRI+CT+IsoSurf: MRI viewer loads up with the CT overlayed on the MRI. 3D figure loads up with the IsoSurface and 3D MRI slices.
Cancel: Abort the process.
Skull Stripping
Skull stripping is done on the MRI volume to get a binary volumetric mask of just the brain region. On applying this mask to volumes like CT, it helps in removing any non-brain tissues like skull, scalp, fat, and any other head tissues from the CT volume. In this tutorial, we have a CT scan of a subject with intracranial electrodes, so on applying this mask not only removes the non-brain tissues but also the hanging extracranial wires. Brainstorm has the following menu options for skull stripping:

SPM: Uses SPM toolbox's Tissue Segmentation.
BrainSuite: Uses BrainSuite's Brain Surface Extractor.
Skip: Proceed without skull stripping.