|
Size: 4198
Comment:
|
Size: 5681
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| <<TableOfContents(2,2)>> | <<TableOfContents(3,2)>> |
| Line 9: | Line 9: |
| You have no data in your database. Some process in the '''Simulate''' menu can generate signals and save them as new files in the database. Because you start from nothing, the Process1 list needs to be empty. Click on Run and try the following processes. | You have no data in your database. The processes in the '''Simulate''' menu can generate signals and save them as new files in the database. Because you start from nothing, the Process1 list needs to be empty. Click on Run and try the following processes. |
| Line 16: | Line 16: |
| === Simulate AR signals === https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity?Simulated_data_.28AR_model.29 |
=== Simulate AR signals [TODO] === For evaluating connectivity metrics, it is useful to create signals that have known connectivity properties. Simulating signals with auto-regressive models for a solution illustrated in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity|Connectivity]]. Two processes are available, using different methods. |
| Line 19: | Line 19: |
| === Simulate AR signals Random === | ==== Process > Simulate AR signals (ARfit) ==== {{attachment:simulate_ar.gif}} ==== Process > Simulate AR signals (random) ==== {{attachment:simulate_ar_random.gif}} |
| Line 25: | Line 30: |
| == Generate full source maps from scout signals == == Simulate MEG/EEG recordings from real sources == |
== Simulate MEG/EEG from sources == |
| Line 32: | Line 36: |
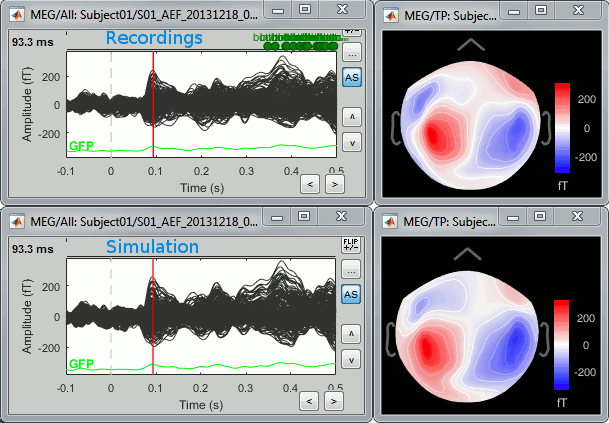
| To simulate MEG/EEG recordings from a minimum norm source model: right-click on the source file, then select the menu '''Model evaluation > Simulate recordings'''. This can be useful for evaluating the quality of the model. The process is documented in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Model_evaluation|Source estimation]]. | ==== From full source maps ==== To simulate MEG/EEG recordings from a minimum norm source model: '''right-click''' on the source file, then select the menu '''Model evaluation > Simulate recordings'''. This can be useful for evaluating the quality of the model. The process is documented in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Model_evaluation|Source estimation]]. |
| Line 36: | Line 41: |
| {{attachment:model_eval_res.gif}} | . {{attachment:model_eval_res.gif||width="378",height="263"}} |
| Line 38: | Line 43: |
| == Simulate MEG/EEG recordings from simulated dipoles == | ==== From scouts ==== If you want to limit this simulation to a specific ROI, one easy solution is to set all the sources outside of the ROI to zero, and calculate the MEG/EEG signal exactly as above. This creates a new file which represents the scalp MEG/EEG recordings if only the selected region of the cortex was activated. Double-click on the source file to open it for display, then in the '''Scout tab''' select the scout(s) of interested and use the menu '''Sources > Simulate recordings'''. {{attachment:simulate_scout.gif}} == Generate full source maps from scouts == You have the values corresponding to source activity of one dipole or a few dipoles, and you would like to save this a source file in the Brainstorm. This source file can later be used to simulate MEG/EEG recordings, exporting as a .nii volume or .gii surface, or any other process in Brainstorm. ==== Constrained sources ==== https://neuroimage.usc.edu/brainstorm/Tutorials/Statistics#Convert_statistic_results_to_regular_files https://neuroimage.usc.edu/brainstorm/Tutorials/TutScouts#Scout_toolbar_and_menus == Simulate MEG/EEG from synthetic dipoles == |
| Line 47: | Line 68: |
| A great simulation article was published by Prof. Debener's neuropsychology lab in Oldenburg. It uses Brainstorm to evaluate and improve the design of ear-EEG devices. All the Matlab/Brainstorm scripts used in this study are available on [[https://figshare.com/articles/software/Matlab-code_for_simulation_of_dipole_sources/11907801|figshare]], together with some videos illustrating the simulations that were done. | A simulation article was published by Prof. Debener's neuropsychology lab in Oldenburg. It uses Brainstorm to evaluate and improve the design of ear-EEG devices. All the Matlab/Brainstorm scripts used in this study are available on [[https://figshare.com/articles/software/Matlab-code_for_simulation_of_dipole_sources/11907801|figshare]], together with some videos illustrating the simulations that were done. |
Simulations
Author: Francois Tadel
This tutorial addresses everything that can be called "simulation" in Brainstorm: simulation of simple signals, simulation of full source maps from a few scouts, simulation of MEG/EEG recordings from source maps (real or synthetic). Its goal is to group all the information that was previously spread across multiple tutorials and forum posts, and difficult to access.
Contents
Simulate signals
You have no data in your database. The processes in the Simulate menu can generate signals and save them as new files in the database. Because you start from nothing, the Process1 list needs to be empty. Click on Run and try the following processes.
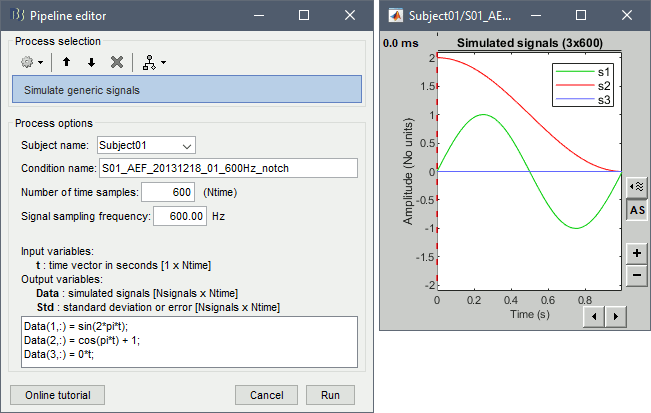
Simulate generic signals
This process allows to define a set of signals with Matlab code and save it in a Brainstorm file. With the text box, you can write a free-form piece of code, which will be evaluated with the function eval(). This code must at least define the variable Data [Nsignals x Ntime], based on in the input time vector t (defined automatically by the process from the number of time points and the sampling frequency). The more rows you add to the Data matrix, the more signals you will have in your file. The example below creates three signals. Another example is available in the tutorial Time-frequency.
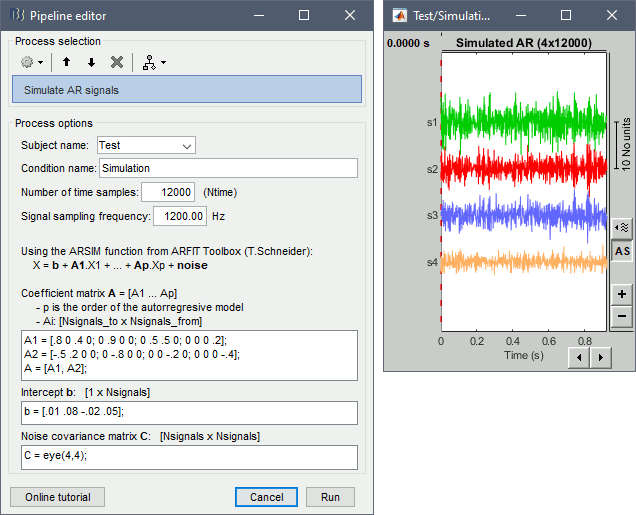
Simulate AR signals [TODO]
For evaluating connectivity metrics, it is useful to create signals that have known connectivity properties. Simulating signals with auto-regressive models for a solution illustrated in the tutorial Connectivity. Two processes are available, using different methods.
Process > Simulate AR signals (ARfit)
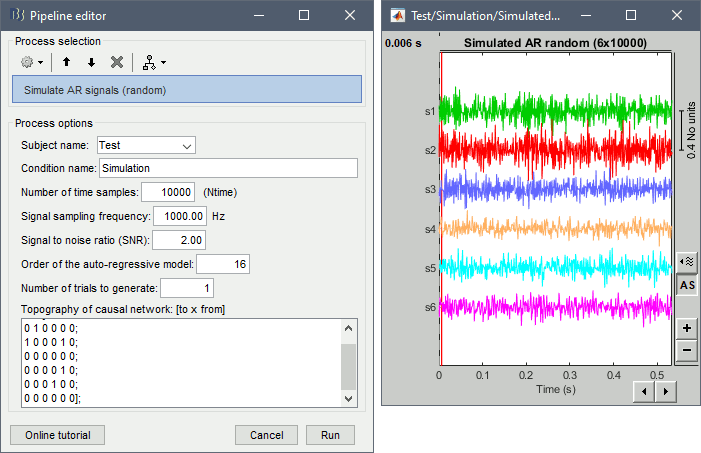
Process > Simulate AR signals (random)
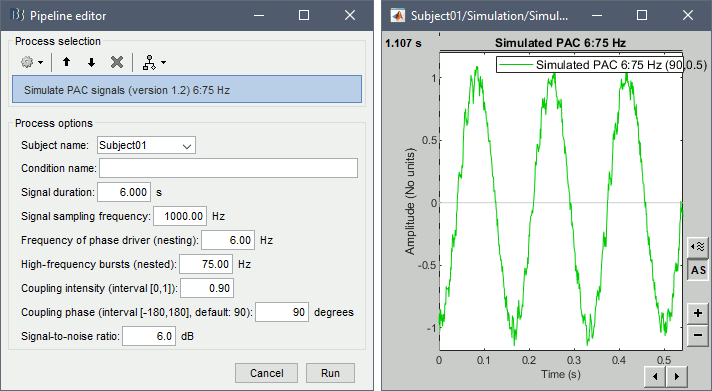
Simulate PAC signals
This process generates synthesized data containing cross-frequency phase-amplitude coupling. It is fully documented in the tutorial Phase-amplitude coupling: Method.
Simulate MEG/EEG from sources
After estimating the sources from real MEG/EEG recordings with the minimum norm method, you can simulate recordings from the model. This is done simply by multiplying the source time series with the forward model:
EEG_sim [Nelec x Ntime] = Forward_model [Nelec x Nsources] * MN_sources [Nsources x Ntime]
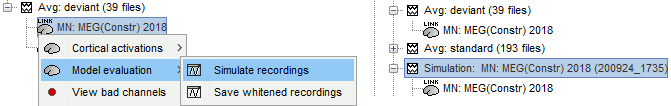
From full source maps
To simulate MEG/EEG recordings from a minimum norm source model: right-click on the source file, then select the menu Model evaluation > Simulate recordings. This can be useful for evaluating the quality of the model. The process is documented in the tutorial Source estimation.
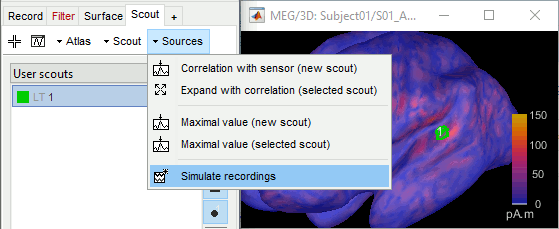
From scouts
If you want to limit this simulation to a specific ROI, one easy solution is to set all the sources outside of the ROI to zero, and calculate the MEG/EEG signal exactly as above. This creates a new file which represents the scalp MEG/EEG recordings if only the selected region of the cortex was activated. Double-click on the source file to open it for display, then in the Scout tab select the scout(s) of interested and use the menu Sources > Simulate recordings.
Generate full source maps from scouts
You have the values corresponding to source activity of one dipole or a few dipoles, and you would like to save this a source file in the Brainstorm. This source file can later be used to simulate MEG/EEG recordings, exporting as a .nii volume or .gii surface, or any other process in Brainstorm.
Constrained sources
https://neuroimage.usc.edu/brainstorm/Tutorials/TutScouts#Scout_toolbar_and_menus
Simulate MEG/EEG from synthetic dipoles
These examples are based on the introduction tutorials, but can easily be transposed to any study. The inputs you need to provide to the process depend on the type of orientation constrains
Constrained
Unconstrained
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_scouts
Additional documentation
Articles
A simulation article was published by Prof. Debener's neuropsychology lab in Oldenburg. It uses Brainstorm to evaluate and improve the design of ear-EEG devices. All the Matlab/Brainstorm scripts used in this study are available on figshare, together with some videos illustrating the simulations that were done.
Meiser A, Tadel F, Debener S, Bleichner MG
The Sensitivity of Ear-EEG: Evaluating the Source-Sensor Relationship Using Forward Modeling
Brain Topography, Aug 2020
Forum discussions
Simulate recordings from sources: http://neuroimage.usc.edu/forums/showthread.php?2563
Simulate recordings from simulated signals: https://neuroimage.usc.edu/forums/t/simulate-scalp-recording/2421/3