|
Size: 16980
Comment:
|
Size: 18488
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 8: | Line 8: |
| <<TableOfContents(2)>> | <<TableOfContents(2,2)>> |
| Line 12: | Line 12: |
| * Go to the [[http://neuroimage.usc.edu/brainstorm3_register/download.php|Download]] page of this website, and dowload the file "bst_sample_mind_neuromag.zip". | * Go to the [[http://neuroimage.usc.edu/brainstorm3_register/download.php|Download]] page of this website and dowload the file "bst_sample_mind_neuromag.zip", unless you already followed the previous tutorial "Review raw recordings and edit markers". |
| Line 15: | Line 15: |
| * In the protocols drop-down list, select the menu "Create new protocol". * Name it "!TutorialNeuromag", and select all the options as illustrated: No default anatomy, Use individual anatomy, Use one channel file per subject (we have only one run, it's possible to share the channels positions and head models between diffrerent conditions).<<BR>><<BR>> {{attachment:createProtocol.gif}} |
* Select the menu File > Create new protocol. Name it "!TutorialNeuromag", and select all the options as illustrated: No default anatomy, Use individual anatomy, Use one channel file per subject (we have only one run, it's possible to share the channels positions and head models between diffrerent conditions).<<BR>><<BR>> {{attachment:createProtocol.gif}} |
| Line 32: | Line 30: |
| * Answer "Yes" to the question "Align surfaces with MRI now?". You should see the following nodes in the database.<<BR>><<BR>> {{attachment:surfListInit.gif}} * "outer skin" = Head, used for surfaces/MRI registration * "inner skull" = Internal surface of the skull, used for the computation of the "overlapping spheres" forward model * "lh" = Left hemisphere * "rh" = Right hemisphere * "pial" = External cortical surface = grey matter/CSF interface (used for source estimation) * "white" = External white matter surface = interface between the grey and the white matter (not used in this tutorial) |
* You should see the following nodes in the database.<<BR>><<BR>> {{attachment:surfListInit.gif}} * '''outer skin''': Head, used for surfaces/MRI registration * '''inner skull''': Internal surface of the skull, used for the computation of the "overlapping spheres" forward model * '''lh''': Left hemisphere * '''rh''': Right hemisphere * '''pial''': External cortical surface = grey matter/CSF interface (used for source estimation) * '''white''': External white matter surface = interface between the grey and the white matter (not used in this tutorial) |
| Line 45: | Line 43: |
| * Select the "Functional data / sorted by subject" exploration mode (second button in the toolbar above the database explorer). | * Select the "Functional data / sorted by subject" exploration mode (second button in the toolbar on top of the database explorer). |
| Line 53: | Line 51: |
| * Click on "STI 001". The full track is read, all the changes of values are extracted and a list of events is created. * Answer "No" when asked whether to save this list of events. This would create a .eve file in the same folder, that would be read instead of the stimulation channel during later access to the raw .fif file. It would provide a much faster access to the events present in the track "STI 001", but we do not need that, because we will have to import the events of the "STI 002". |
* Select "STI 001" and "STI 002". The full track is read, all the changes of values are detected from those two tracks and a list of events is created. |
| Line 56: | Line 53: |
| * __Time window__: Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations. * __Split__: Useful to import continuous recordings without events. We do not need this here. * __Events selection__: Check the "Use events" option, and select "Event #5". * The value between parenthesis represents the number of occurrences of this event. * Event #5 represents the instants where the value of track STI 001 changing from 0 to 5 (electric stimulation) * Event #0 is set when the value goes back to zero * __Epoch time__: Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms * __Use Signal Space Projections__: Save and use the SSP vectors created by !MaxFilter or during other pre-processing steps. Keep this option selected. * __Remove DC Offset__: Check this option, and select: Time range: [-100, 0] ms. <<BR>>For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), and subtract it from the channel at all the times in [-100,+300]ms. * __Resample recordings__: Keep this unchecked * __Create new conditions for epochs__: If selected, a new condition is created for each event type (here, it will create a condition "Event_5"). If not selected, all the epochs from all the selected events are saved in the same condition, the one that was selected in the database explorer (if no condition selected, create a condition called "New condition"). |
* '''Time window''': Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations. * '''Split''': Useful to import continuous recordings without events. We do not need this here. * '''Events selection''': Check the "Use events" option, and select both STI001 and STI002. The value between parenthesis represents the number of occurrences of this event. * '''Epoch time''': Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms * '''Use Signal Space Projections''': Save and use the SSP vectors created by !MaxFilter or during other pre-processing steps. Keep this option selected. * '''Remove DC Offset''': Check this option, and select: Time range: [-100, 0] ms. <<BR>>For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), and subtract it from the channel at all the times in [-100,+300]ms. * '''Resample recordings''': Keep this unchecked * '''Create new conditions for epochs''': If selected, a new condition is created for each event type (here, it will create a condition "Event_5"). If not selected, all the epochs from all the selected events are saved in the same condition, the one that was selected in the database explorer (if no condition selected, create a condition called "New condition"). |
| Line 68: | Line 62: |
| * At the end, you're asked if you want to "Refine the registration with the head points". This operation improves the initial MRI/MEG registration by applying and ICP algorithm to make fit the head points digitized at the MEG acquisition and the scalp surface. Answer yes. You'll see that, even if the result is not perfect, it dramatically improves the positioning of the head in the MEG helmet.<<BR>><<BR>> {{attachment:refine.gif}} | |
| Line 70: | Line 65: |
| * Rename the condition "Event_5" into "Right" (Right-click > Rename, or press F2, or click twice waiting a while before the two clicks). * Repeat all those steps to import the events from track "STI 002". Rename "Event_5" into "Left". Your database should look like this:<<BR>><<BR>> {{attachment:treeAfterImport2.gif}} * __Note__: if you do not rename the condition "Event_5" in "Right" between the two imports, you would have at the end all the epochs in the same Event_5 condition. In this case, another way (and maybe safer) way to proceed is to: * Create two conditions Left and Right before starting to import the recordings. * Import the "STI 001" events in condition Right, and the "STI 002" in Left (do not forget to uncheck the option "Create new conditions for epochs", if not the condition selection would be overridden) |
* Rename the condition "STI_001_5" and "STI_002_5" into respectively "Right" and "Left" (Right-click > Rename, or press F2, or click twice waiting a while before the two clicks).<<BR>><<BR>> {{attachment:treeAfterImport2.gif}} * __Note__: Another way to proceed would be: 1) to create a new empty condition Right, and 2) inside it import the STI001 events only. Same for Left and STI002. It can be a safer way to proceed, depending on the cases. |
| Line 77: | Line 69: |
| === Stimulation artifact removal === The electric stimulation of the median nerve induces a strong artefact right after 0ms. We are going to use a simple trick to remove this artifact: re-interpolate the values between 0ms and 4ms (linear interpolation). It doesn't affect much the data but will make all the displays much better. |
=== Select the files to process === We want to apply the same pre-processing operations to all the trials of both conditions. Drag'n'drop the "Subject01" node from the database explorer to the tab ''Process1''. Select on the "Recordings" button, then click ''Run''. |
| Line 80: | Line 72: |
| * Drag'n'drop the "Subject01" node from the database explorer to the "Processes" tab. Click on the "Recordings" button, and then click on "run process".<<BR>><<BR>> {{attachment:cutStimList.gif}} * Select "Process > Cut stimulation artifact". The baseline time windows represents here the time instants for which the values are going to be re-interpolated. Set it to [0, 3.9]ms. <<BR>><<BR>> {{attachment:cutStimOptions.gif}} * Click on Run. All the files are going to be replaced with the processed ones. |
. {{attachment:cutStimList.gif}} |
| Line 84: | Line 74: |
| === Band-pass filtering === * Drag'n'drop again the "Subject01" node in the list of files to be processed. Select "Process > Band-pass filtering". Select the option "Overwrite intial files". Click on Run. |
=== Create a processing pipeline === * '''Process > Cut stimulation artifact''': The electric stimulation of the median nerve induces a strong artefact right after 0ms. This process uses a simple trick to remove this artifact: re-interpolate the values for the artifact time window (linear interpolation). It doesn't affect much the data but will make all the displays much better. Set the time window to [0, 3.9]ms. Select "Overwrite initial file". |
| Line 87: | Line 77: |
| * Set the frequency range to [0, 90] Hz.<<BR>><<BR>> {{attachment:bandpassSelection.gif}} * With all those process tags added, the comments of the files are getting a little too long. Rename them respectively Right and Left, by renaming directly the list (the node that contains all the epochs in each condition). It takes a while but will improve a lot the readability later.<<BR>><<BR>> {{attachment:treeAfterPreprocess.gif}} |
* '''Process > Band-pass filter''': Set the frequency range to [2Hz -90Hz]. Select "Overwrite initial file". |
| Line 90: | Line 79: |
| === Review the epochs === It is always very important to keep an eye on the quality of the data at the different steps of the analysis. There is always a few epochs that are too artifacted or noisy to be used, or one bad sensor. Unfortunately, there are no tools yet in Brainstorm for autoamtic artifact detection and rejection. But here is a procedure for reviewing your epochs manually. |
* '''Detect > Detect bad channels: peak-to-peak''': This process tests the peak-to-peak amplitude of each channel, and defines it is good or bad based on a threshold value that is defined individually for each channel type.Set the following options: * '''Time window''': All the epoch (keep the default value) * '''Meg gradio''': [0, 3500] * '''Meg magneto''': [0, 3500] * '''Reject entire trial''' * '''Average > Average files''': Select "By condition".<<BR>><<BR>> {{attachment:pipeline.gif}} === Run the pipeline === Click on run. Wait. With all those process tags added, the comments of the files are getting a little too long. Rename them respectively Right and Left, by renaming directly the list (the node that contains all the epochs in each condition). It takes a while but will improve a lot the readability later. . {{attachment:treeAfterPreprocess.gif}} {{attachment:treeAfterPreprocess2.gif}} Expand the list of trials Left, and notice that the icons of a few trials have a red flag. * Those are the trials that were identified as bad by the process "Detect bad channels". * Now if you clear the Process1 list and drag and drop again Subject01, you would see the following: there are now only 598 recording files detected, instead of 624 previously. This number is: number total of trials (624) - number of bad trials (28) + the averages (2). * On the right side of the panel, a list of checkboxes appeared. They represent the list of tags that were found in the collection of files in Subject01. Tags "bandpass" and "cutstim" are set for all the trials, and the "none" check box represents the files that do not have tags (the two averages). * If you uncheck "none", it removes the 2 averaged files from the selection. If you uncheck both "bandpass" and "cutstim", it removes all the pre-processed trials from the selection. . {{attachment:treeBad.gif}} {{attachment:treeBadProcess.gif}} === Review the epochs manually === It is always very important to keep an eye on the quality of the data at the different steps of the analysis. There are always a few epochs that are too artifacted or noisy to be used, or one bad sensor. An automatic detection was already applied, but here is the procedure if you need to do it manually. |
| Line 96: | Line 109: |
| * You can tag some channels as bad: select a sensor by clicking on it and press the Delete key (or right-click > Channel > Mark selected as bad) * You can completely delete an epoch if you consider it as unusable (delete the files from the tree). |
* '''Tag channels as bad''': select a sensor by clicking on it and press the Delete key (or right-click > Channel > Mark selected as bad) * '''Tag the entire trial as bad''': You can mark a trial as bad, it will be completely ignored from all the following computations. Right-click 1) on the trial in the explorer or 2) in the time series figure, and select menu ''Reject trial''. Or use the keyboard shortcut ''Ctrl+B''. |
| Line 99: | Line 112: |
| * Do the same for the Left condition. | |
| Line 101: | Line 113: |
| === Averaging === You can use Brainstorm to work on individual trials or on average recordings. But even if you plan to work on single trials, start your exploration of the recordings by computing an average per condition. It would give you a good idea of the quality of your recordings and pre-processing operations. If you do not see anything looking like the effect you are supposed to observe on the average, it is a complete waste of time to go on with source or time-frequency analysis. * Drag'n'drop again the node "Subject01" in the Processes tab, and click on Run process. * Select "Average > Average by condition (Grand average). Click on Run. * Two new files called "GAVE: ...." appeared in the database. They are the averaged response for each condition, locked on the stimulus.<<BR>><<BR>> {{attachment:treeAfterAverage.gif}} |
=== Review the average === |
| Line 117: | Line 124: |
| * Right-click on the channel file > MRI Registration > Check MEG-MRI registration<<BR>><<BR>> {{attachment:checkRegistration.gif}} {{attachment:checkRegistrationFig.gif}} * The yellow squares represent the Neuromag MEG sensors, and the green dots represent the head points that where saved with the MEG recordings. * In this case the registration looks acceptable. If you consider it is not your case, you can try the two menus: * Auto-registration: This operation will try to find a better registration of the head points (green dots), on the head surface (grey surface) * Manual resgistration: You rotate/translate the sensor array manually respect to the head surface. |
* Right-click on the channel file > MRI Registration > Check<<BR>><<BR>> {{attachment:checkRegistration.gif}} {{attachment:checkRegistrationFig.gif}} * The yellow surface represents the inner surface of the MEG helmet, and the green dots represent the head points that where saved with the MEG recordings. * In this case the registration looks acceptable. If you consider it is not your case, you can try the two other menus: "Edit..." and "Refine using head points" |
| Line 125: | Line 130: |
| * Select "overlapping spheres" and click on Run. Select "Inner skull" when asked what surface to use to estimate the spheres. For more information, read the tutorial "[[Tutorials/TutHeadModel|Computing a head model]]".<<BR>><<BR>> {{attachment:headmodelSurface.gif}} | |
| Line 132: | Line 136: |
| * Right-click on one of them and select: Noise covariance > Compute from recordings. Leave all the options unchanged, and click on Ok.<<BR>><<BR>> {{attachment:noisecovMenu.gif}} {{attachment:noisecovOptions.gif}} * This operation computes the noise covariance matrix based on the baseline, [-100, 0]ms, of all the trials (624 files). The resulting file is also stored in the "Common files" node. |
* Right-click on one of them and select: Noise covariance > Compute from recordings. Leave all the options unchanged, and click on Ok. * This operation computes the noise covariance matrix based on the baseline, [-100, 0]ms, of all the __good__ trials (596 files). The result is stored in a new file in the "Common files" folder. . {{attachment:noisecovMenu.gif}} {{attachment:noisecovOptions.gif}} == Source estimation == Reconstruction of the cortical sources with a weighted minimum norm estimator (wMNE). * Right-click on "Common files", (or on the head model, or on the subject node, this is all equivalent), and select "Compute sources". <<BR>><<BR>> {{attachment:inverseMenu.gif}} * Select "Yes" to ignore the warning about noise covariance used at the same time for averages and single trials (if you do not see such a warning, that's fine too). In this case, this warning does not really apply, the results are the same if we compute separately the minimum norm estimator for the single trials and the averages. * Select "Normal mode" (not "Expert mode"), wMNE, and both gradiometers and magnetometers, as in the figure below. Click on Run.<<BR>><<BR>> {{attachment:inverseOptions.gif}} {{attachment:treeAfterInverse.gif}} * A shared inversion kernel was created in the ''(Common files)'' folder, and a link node is now visible for each recording file, single trials and averages. For more information about what those links mean and the operations performed to display them, please refer to the [[Tutorials/TutSourceEstimation|tutorial #4 "Source estimation"]]. * For the average of condition Right: display the time series, the 2D sensor topography, the sources on the cortex, the sources on 3D orthogonal slices, and the sources in the MRI Viewer.<<BR>><<BR>> {{attachment:inverseAllDisplay.gif}} * Create a scout called LeftSS that covers the most active region of the cortex at 33ms for condition Right. Display its time series for conditions Left and Right at the same time (select both Left and Right source files, right click > Cortical activations > Scouts time series (check "Overlay: conditions").<<BR>><<BR>> {{attachment:inverseScoutLeft.gif}} == Time-frequency == |
Import and process Neuromag raw recordings
This tutorial describes how to process raw Neuromag recordings. It is based on median nerve stimulation acquired at MGH in 2005 with a Neuromag Vectorview 306 system. The sample dataset contains the results for one subject for both arms.
This document shows what to do step by step, but does not really explain what is happening, the meaning of the different options or processes, the issues or bugs you can encounter, and does not provide an exhaustive description of the software features. Those topics are introduced in the basic tutorials based on CTF recordings; so make sure that you followed all those initial tutorials before going through this one.
The script file tutorial_mind_neuromag.m in the brainstorm3/toolbox/script folder performs exactly the same tasks automatically, without any user interaction. Please have a look at this file if you plan to write scripts to process recordings in .fif format.
Contents
Download and installation
- It is considered that you followed all the basic tutorials and that you already have a fully working copy of Brainstorm installed on your computer.
Go to the Download page of this website and dowload the file "bst_sample_mind_neuromag.zip", unless you already followed the previous tutorial "Review raw recordings and edit markers".
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself.
- Start Brainstorm (Matlab scripts or stand-alone version)
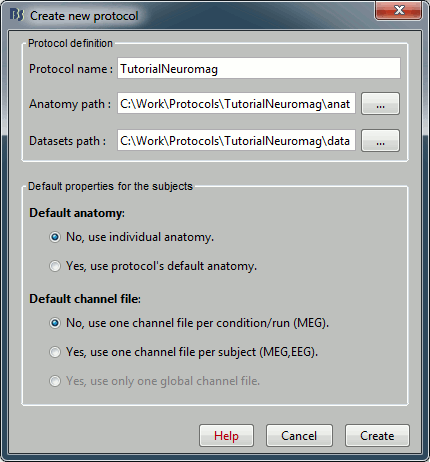
Select the menu File > Create new protocol. Name it "TutorialNeuromag", and select all the options as illustrated: No default anatomy, Use individual anatomy, Use one channel file per subject (we have only one run, it's possible to share the channels positions and head models between diffrerent conditions).
Importing anatomy
Create the subject
- Select the "Anatomy" exploration mode (first button in the toolbar above the database explorer).
Right-click on the protocol node and select "New subject".
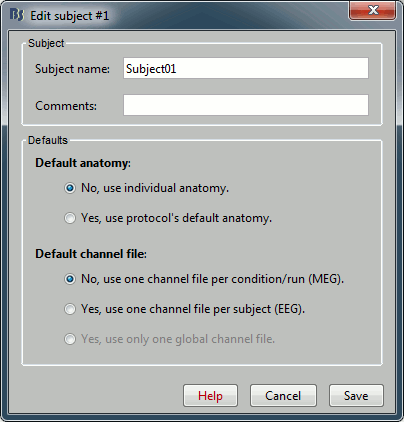
You can leave the default name "Subject01" and the default options. Then click on Save.
Import the MRI
Right-click on the subject node, and select "Import MRI". Select the file: sample_mind_neuromag/anatomy/T1.mri
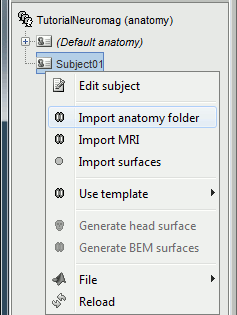
The orientation of the MRI is already ok, you just have to mark all the six fiducial points, as explained in the following wiki page: Coordinate systems. Click on save button when you're done.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
If at some point you do not see the files you've just imported in the database explorer, you just need to refresh the display: by pressing F5, or using the menu File > Refresh tree display.
Import the surfaces
Right-click again on the subject node, and select "Import surfaces". Select the "FreeSurfer surfaces" file type, and select all the surface files at once, holding the SHIFT or CTRL buttons. Click on Open.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
You should see the following nodes in the database.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
outer skin: Head, used for surfaces/MRI registration
inner skull: Internal surface of the skull, used for the computation of the "overlapping spheres" forward model
lh: Left hemisphere
rh: Right hemisphere
pial: External cortical surface = grey matter/CSF interface (used for source estimation)
white: External white matter surface = interface between the grey and the white matter (not used in this tutorial)
Downsample the two pial and the two white surfaces to 7,500 vertices each (right-click > Less vertices)
Merge the two downsampled hemispheres of the pial surface (select both lh.pial_7500 and rh.pial_7500 files, right-click > Merge surfaces). Rename the new surface in "cortex".
- Do the same with the white matter. Call the result "white".
Delete all the intermediate lh and rh surfaces. Rename the head and the inner skull with shorter names. Display all the surfaces and play with the colors and transparency to check that everything what imported correctly. You should obtain something like this:
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Importing MEG recordings
- Select the "Functional data / sorted by subject" exploration mode (second button in the toolbar on top of the database explorer).
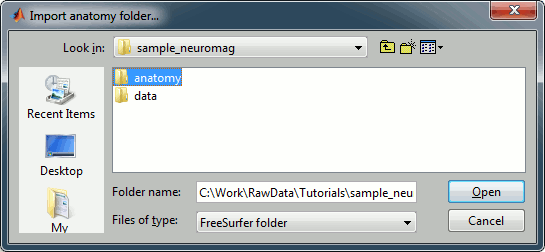
Right click on the subject, select Import MEG/EEG. Select the "Neuromag FIFF" file type, and open the only .fif file in the folder sample_mind_neuromag/data.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Select "Event channel" when asked about the events.
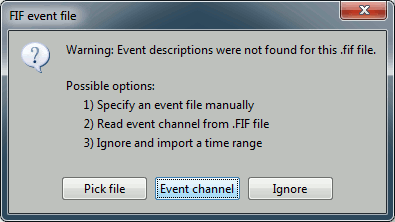
You have then to select from which technical track you want to read the events.
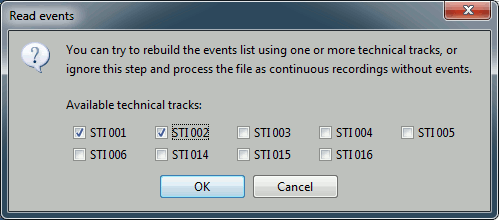
- In this dataset, the tracks of interest are:
STI 001: Contains the onsets of the electric stimulation of the right arm.
STI 002: Contains the onsets of the electric stimulation of the left arm.
- Those are not standard settings, they depend on your acquisition setup. In most cases, you would rather import "events" defined with the Neuromag acquisition software, which are saved in tracks STI 014, STI 101, STI 201...
- Select "STI 001" and "STI 002". The full track is read, all the changes of values are detected from those two tracks and a list of events is created.
The following figure appears, that asks how to import those recordings in the Brainstorm database.
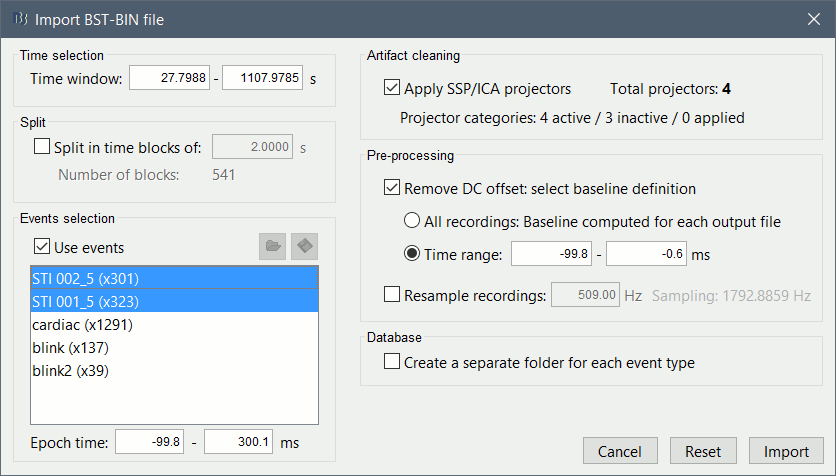
Time window: Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations.
Split: Useful to import continuous recordings without events. We do not need this here.
Events selection: Check the "Use events" option, and select both STI001 and STI002. The value between parenthesis represents the number of occurrences of this event.
Epoch time: Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms
Use Signal Space Projections: Save and use the SSP vectors created by MaxFilter or during other pre-processing steps. Keep this option selected.
Remove DC Offset: Check this option, and select: Time range: [-100, 0] ms.
For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), and subtract it from the channel at all the times in [-100,+300]ms.Resample recordings: Keep this unchecked
Create new conditions for epochs: If selected, a new condition is created for each event type (here, it will create a condition "Event_5"). If not selected, all the epochs from all the selected events are saved in the same condition, the one that was selected in the database explorer (if no condition selected, create a condition called "New condition").
- Click on Import and wait.
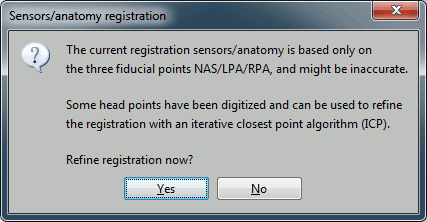
At the end, you're asked if you want to "Refine the registration with the head points". This operation improves the initial MRI/MEG registration by applying and ICP algorithm to make fit the head points digitized at the MEG acquisition and the scalp surface. Answer yes. You'll see that, even if the result is not perfect, it dramatically improves the positioning of the head in the MEG helmet.
The files that appear in the folder "(Common files)" will be shared for all the conditions of Subject01. This mechanism allows to compute the forward and inverse models only once for all the conditions when there is only one run (= one channel file = one position of sensors recorded).
Rename the condition "STI_001_5" and "STI_002_5" into respectively "Right" and "Left" (Right-click > Rename, or press F2, or click twice waiting a while before the two clicks).
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Note: Another way to proceed would be: 1) to create a new empty condition Right, and 2) inside it import the STI001 events only. Same for Left and STI002. It can be a safer way to proceed, depending on the cases.
Pre-processing
Select the files to process
We want to apply the same pre-processing operations to all the trials of both conditions. Drag'n'drop the "Subject01" node from the database explorer to the tab Process1. Select on the "Recordings" button, then click Run.
Create a processing pipeline
Process > Cut stimulation artifact: The electric stimulation of the median nerve induces a strong artefact right after 0ms. This process uses a simple trick to remove this artifact: re-interpolate the values for the artifact time window (linear interpolation). It doesn't affect much the data but will make all the displays much better. Set the time window to [0, 3.9]ms. Select "Overwrite initial file".
Process > Band-pass filter: Set the frequency range to [2Hz -90Hz]. Select "Overwrite initial file".
Detect > Detect bad channels: peak-to-peak: This process tests the peak-to-peak amplitude of each channel, and defines it is good or bad based on a threshold value that is defined individually for each channel type.Set the following options:
Time window: All the epoch (keep the default value)
Meg gradio: [0, 3500]
Meg magneto: [0, 3500]
Reject entire trial
Run the pipeline
Click on run. Wait. With all those process tags added, the comments of the files are getting a little too long. Rename them respectively Right and Left, by renaming directly the list (the node that contains all the epochs in each condition). It takes a while but will improve a lot the readability later.
Expand the list of trials Left, and notice that the icons of a few trials have a red flag.
- Those are the trials that were identified as bad by the process "Detect bad channels".
- Now if you clear the Process1 list and drag and drop again Subject01, you would see the following: there are now only 598 recording files detected, instead of 624 previously. This number is: number total of trials (624) - number of bad trials (28) + the averages (2).
- On the right side of the panel, a list of checkboxes appeared. They represent the list of tags that were found in the collection of files in Subject01. Tags "bandpass" and "cutstim" are set for all the trials, and the "none" check box represents the files that do not have tags (the two averages).
- If you uncheck "none", it removes the 2 averaged files from the selection. If you uncheck both "bandpass" and "cutstim", it removes all the pre-processed trials from the selection.
Review the epochs manually
It is always very important to keep an eye on the quality of the data at the different steps of the analysis. There are always a few epochs that are too artifacted or noisy to be used, or one bad sensor. An automatic detection was already applied, but here is the procedure if you need to do it manually.
Double-click on the Right epochs list to expand the list of files contained in it. Then display the time series for the first Right epoch by double-clicking on it.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Then press F3 to go to the next epoch (or menu "Navigator > Next data file" in the main Brainstorm window). Do that until you reach the last epoch. For each epoch:
- Check that the amplitude range in consistant with the previous epochs (in this study: between 1000fT and 2500fT).
Tag channels as bad: select a sensor by clicking on it and press the Delete key (or right-click > Channel > Mark selected as bad)
Tag the entire trial as bad: You can mark a trial as bad, it will be completely ignored from all the following computations. Right-click 1) on the trial in the explorer or 2) in the time series figure, and select menu Reject trial. Or use the keyboard shortcut Ctrl+B.
- In the case of this study, there are so many trials for each condition and the recorded signal is so strong that you do not really need to spend too much time on the selection of the bad channels and bad epochs. If you just keep everything as it is, it is just fine.
Review the average
Display the time series for each average: Right-click on the file > MEG (All) > Display time series.
Display the 2D topographies for each average: Right-click on the file > MEG (All) > 2D Sensor cap (or press CTRL+T from the time series figures)
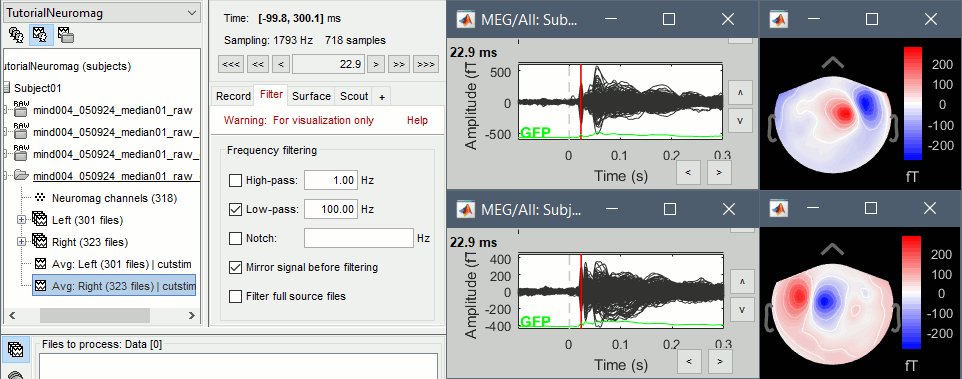
- Everything looks ok, the time series figures show a clean signal with a very high SNR, and the topographies at the first response peak (about 22ms) show an activity on the left side for the Right condition, and on the right side for the Left condition.
Forward model
First step of the source estimation process: establishing a model that describes the way the brain electric activities influence the magnetic fields that are recorded by the MEG sensors. This model can be designated in the documentation by the following terms: head model, forward model, lead field matrix.
MEG / MRI registration
An accurate forward model requires first of all an accurate registration of the anatomy files (MRI+surfaces) and functional recordings (position of the MEG sensors and EEG electrodes). A basic registration is provided by the alignment of the fiducials (Nasion, LPA, RPA), that were both located before the acquisition of the recordings and marked on the MRI in Brainstorm. This registration based on three points only can be very inaccurate, because it is sometimes hard to identify clearly those points, and not everybody identify them the same way. Two methods described in the ?introduction tutorial #3 may help you to improve this registration.
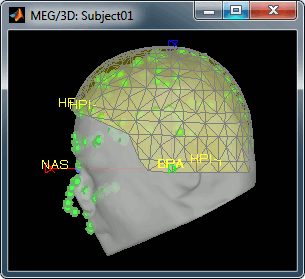
- The yellow surface represents the inner surface of the MEG helmet, and the green dots represent the head points that where saved with the MEG recordings.
- In this case the registration looks acceptable. If you consider it is not your case, you can try the two other menus: "Edit..." and "Refine using head points"
Compute head model
Right-click on any node that contains the channel file (including the channel file itself), and select: "Compute head model".
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
- When the computation is done, close the "Check spheres" figure. The lead field matrix is saved in file "Overlapping spheres" in "Common files", in a the "Gain" field.
Noise covariance matrix
To estimate the sources properly, we need an estimation of the noise level for each sensor. A good way to do this is to compute the covariance matrix of the concatenation of the baselines from all the trials in both conditions.
- Select at the same time the two groups of trials (Right and Left). To do this: hold the Control key and click successively on the Right and the Left trial lists.
Right-click on one of them and select: Noise covariance > Compute from recordings. Leave all the options unchanged, and click on Ok.
This operation computes the noise covariance matrix based on the baseline, [-100, 0]ms, of all the good trials (596 files). The result is stored in a new file in the "Common files" folder.
Source estimation
Reconstruction of the cortical sources with a weighted minimum norm estimator (wMNE).
Right-click on "Common files", (or on the head model, or on the subject node, this is all equivalent), and select "Compute sources".
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
- Select "Yes" to ignore the warning about noise covariance used at the same time for averages and single trials (if you do not see such a warning, that's fine too). In this case, this warning does not really apply, the results are the same if we compute separately the minimum norm estimator for the single trials and the averages.
Select "Normal mode" (not "Expert mode"), wMNE, and both gradiometers and magnetometers, as in the figure below. Click on Run.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
A shared inversion kernel was created in the (Common files) folder, and a link node is now visible for each recording file, single trials and averages. For more information about what those links mean and the operations performed to display them, please refer to the ?tutorial #4 "Source estimation".
For the average of condition Right: display the time series, the 2D sensor topography, the sources on the cortex, the sources on 3D orthogonal slices, and the sources in the MRI Viewer.
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)
Create a scout called LeftSS that covers the most active region of the cortex at 33ms for condition Right. Display its time series for conditions Left and Right at the same time (select both Left and Right source files, right click > Cortical activations > Scouts time series (check "Overlay: conditions").
![[ATTACH] [ATTACH]](/moin_static198/brainstorm1/img/attach.png)