|
Size: 1944
Comment:
|
Size: 5113
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 14: | Line 14: |
| You can follow this tutorial after processing the recordings for the 19 subjects (6 runs per subject) as illustrated in the [[Tutorials/VisualSingle|single subject tutorial]]. Otherwise, we provide a Brainstorm protocol that includes all the imported data, downsampled at 275Hz. | You can follow this tutorial after processing the recordings for the 19 subjects (6 runs per subject) as illustrated in the [[Tutorials/VisualSingle|single subject tutorial]]. Otherwise, we provide a Brainstorm protocol that includes all the imported data, downsampled at 275Hz: |
| Line 20: | Line 20: |
| == Topics to include == * Process: Sources > Spatial smoothing |
The database you need in order to follow this tutorial should contain the following: |
| Line 23: | Line 22: |
| * The individual anatomy imported from FreeSurfer or BrainSuite for each subject (19 subjects). * The sensor level averages (MEG+EEG) for each subject and each run (downsampled to 275Hz or not). * The forward model, noise covariance and inverse models for each subject and each run. <<BR>><<BR>> {{attachment:db_tutorialgroup.gif||height="346",width="422"}} == Overview of the analysis == === Coregistration of the acquisition runs === For each subject, all the runs have been registered to a common head position with MaxFilter. To verify this, select all the channel files within one subject, right-click > Display sensors > MEG (all). The surfaces representing the MEG helmet are perfectly overlapping for all the runs. When the runs are not aligned, it looks like [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Multiple_runs_and_head_positions|this]]. . {{attachment:run_coreg.gif||height="163",width="486"}} This means that we can safely average or compare the MEG sensor values across runs within one subject. However, it is not reliable to average MEG recordings across subjects, because of the [[http://neuroimage.usc.edu/brainstorm/Tutorials/Workflows#MEG_recordings|anatomical differences between subjects]]. This doesn't mean that we can estimate the sources only once per subject. We have computed different SSP projectors and different bad channels for the each acquisition run. To be able to use this information efficiently we should estimate the sources for the trial averages for each run separately, then average the sources across runs. The forward model is the same for all the runs within one subject, therefore it can be computed for the first run and copied to all the other runs. === Objectives [TODO] === The objectives for this tutorial are to reproduce the analysis presented in .... * 1 * 2 * 3 The methodology that we will follow for computing the averages and the other statistics are described in the tutorial "[[Tutorials/Workflows|Workflows]]". == Bad subjects == After evaluating the results for the 19 subjects, 3 subjects were excluded from the analysis: * '''sub001''': Error during the digitization of the EEG electrodes with the Polhemus (the same electrode was clicked twice). This impacts only the EEG, this subject could be used for MEG analysis. <<BR>><<BR>> {{attachment:bad_sub001.gif||height="158",width="274"}} * '''sub005''': Too many blinks or other eye movements. <<BR>> <<BR>> {{attachment:bad_sub005.gif||height="164",width="430"}} * '''sub016''': Too many blinks or other eye movements. <<BR>> <<BR>> {{attachment:bad_sub016.jpg||height="152",width="430"}} To ignore these three subjects from the group analysis without having to delete them from the database, is to mark all the recordings they contain as "bad". * In the database explorer, select the the three averages in '''sub001/run_01_sss_notch'''. * Right-click on one one them > Reject trials. * Repeat the operation for all the runs of the three subjects sub001, sub005, sub016. <<BR>><<BR>> {{attachment:bad_subjects.gif}} == Subject and group averages == |
MEG visual tutorial: Group analysis
Authors: Francois Tadel, Elizabeth Bock.
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "Multimodal, Multisubject data fusion". The data processed here consists in simultaneous MEG/EEG recordings of 19 subjects performing simple visual task on a large number of famous, unfamiliar and scrambled faces. This tutorial follows another page that explains how to process one single subject in details.
Contents
License
This dataset was obtained from the OpenfMRI project (http://www.openfmri.org), accession #ds117. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset, Scientific Data (2015)
Any questions, please contact: rik.henson@mrc-cbu.cam.ac.uk
Download and installation
You can follow this tutorial after processing the recordings for the 19 subjects (6 runs per subject) as illustrated in the single subject tutorial. Otherwise, we provide a Brainstorm protocol that includes all the imported data, downsampled at 275Hz:
Go to the Download page, download the file TutorialGroup.zip.
- Unzip this file in your Brainstorm database folder (brainstorm_db).
In Brainstorm, menu File > Load protocol > Load from folder > Select brainstorm_db/TutorialGroup
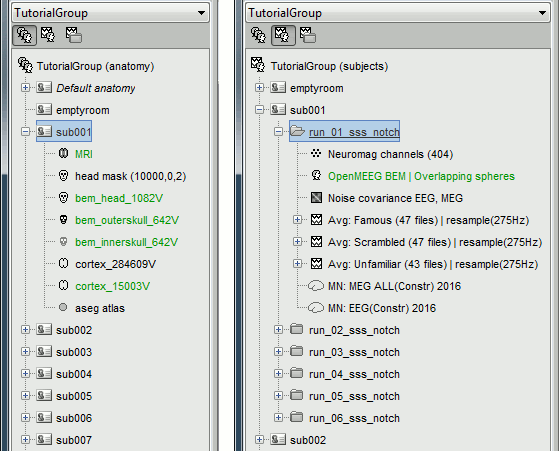
The database you need in order to follow this tutorial should contain the following:
The individual anatomy imported from FreeSurfer or BrainSuite for each subject (19 subjects).
- The sensor level averages (MEG+EEG) for each subject and each run (downsampled to 275Hz or not).
The forward model, noise covariance and inverse models for each subject and each run.
Overview of the analysis
Coregistration of the acquisition runs
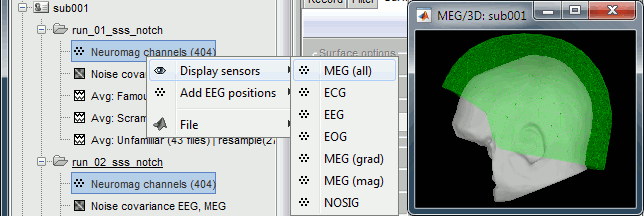
For each subject, all the runs have been registered to a common head position with MaxFilter. To verify this, select all the channel files within one subject, right-click > Display sensors > MEG (all). The surfaces representing the MEG helmet are perfectly overlapping for all the runs. When the runs are not aligned, it looks like this.
This means that we can safely average or compare the MEG sensor values across runs within one subject. However, it is not reliable to average MEG recordings across subjects, because of the anatomical differences between subjects.
This doesn't mean that we can estimate the sources only once per subject. We have computed different SSP projectors and different bad channels for the each acquisition run. To be able to use this information efficiently we should estimate the sources for the trial averages for each run separately, then average the sources across runs.
The forward model is the same for all the runs within one subject, therefore it can be computed for the first run and copied to all the other runs.
Objectives [TODO]
The objectives for this tutorial are to reproduce the analysis presented in ....
- 1
- 2
- 3
The methodology that we will follow for computing the averages and the other statistics are described in the tutorial "Workflows".
Bad subjects
After evaluating the results for the 19 subjects, 3 subjects were excluded from the analysis:
sub001: Error during the digitization of the EEG electrodes with the Polhemus (the same electrode was clicked twice). This impacts only the EEG, this subject could be used for MEG analysis.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
To ignore these three subjects from the group analysis without having to delete them from the database, is to mark all the recordings they contain as "bad".
In the database explorer, select the the three averages in sub001/run_01_sss_notch.
Right-click on one one them > Reject trials.
Repeat the operation for all the runs of the three subjects sub001, sub005, sub016.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Subject and group averages