|
Size: 6514
Comment:
|
Size: 35648
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = MEG visual tutorial: Single subject = | = MEG visual tutorial: Single subject (BIDS) = |
| Line 4: | Line 4: |
| The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "[[ftp://ftp.mrc-cbu.cam.ac.uk/personal/rik.henson/wakemandg_hensonrn/Publications/SPM12_manual_chapter.pdf|Multimodal, Multisubject data fusion]]". The data processed here consists in simulateneous MEG/EEG recordings of 16 subjects performing simple visual task on a large number of famous, unfamiliar and scrambled faces. The analysis is split in two tutorial pages: the present tutorial describes the detailed analysis of one single subject and another one that the describes the batch processing and [[Tutorials/VisualGroup|group analysis of the 16 subjects]]. Note that the operations used here are not detailed, the goal of this tutorial is not to teach Brainstorm to a new inexperienced user. For in depth explanations of the interface and the theory, please refer to the introduction tutorials. |
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "[[ftp://ftp.mrc-cbu.cam.ac.uk/personal/rik.henson/wakemandg_hensonrn/Publications/SPM12_manual_chapter.pdf|Multimodal, Multisubject data fusion]]". We use here a recent update of this dataset, reformatted to follow the [[http://bids.neuroimaging.io|Brain Imaging Data Structure (BIDS)]], a standard for neuroimaging data organization. It is part of a collective effort to document and standardize MEG/EEG group analysis, see Frontier's research topic: [[https://www.frontiersin.org/research-topics/5158|From raw MEG/EEG to publication: how to perform MEG/EEG group analysis with free academic software]]. The data processed here consists of simultaneous MEG/EEG recordings from 16 participants performing a simple visual recognition task from presentations of famous, unfamiliar and scrambled faces. The analysis is split in two tutorial pages: the present tutorial describes the detailed interactive analysis of one single subject; the second tutorial describes batch processing and [[Tutorials/VisualGroup|group analysis of all 16 participants]]. Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the [[http://neuroimage.usc.edu/brainstorm/Tutorials#Get_started|introduction tutorials]]. |
| Line 13: | Line 13: |
| These data are provided freely for research purposes only (as part of their Award of the BioMag2010 Data Competition). If you wish to publish any of these data, please acknowledge Daniel Wakeman and Richard Henson. The best single reference is: Wakeman DG, Henson RN, [[http://www.nature.com/articles/sdata20151|A multi-subject, multi-modal human neuroimaging dataset]], Scientific Data (2015) Any questions, please contact: rik.henson@mrc-cbu.cam.ac.uk |
This dataset was obtained from the OpenNeuro project ([[https://openneuro.org/|http://openneuro.org]]), accession [[https://openneuro.org/datasets/ds000117|#ds117]]. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data: Wakeman DG, Henson RN, [[http://www.nature.com/articles/sdata20151|A multi-subject, multi-modal human neuroimaging dataset]], Scientific Data (2015) Any questions regarding the data, please contact: rik.henson@mrc-cbu.cam.ac.uk |
| Line 19: | Line 19: |
| * 16 subjects * 6 runs (sessions) of approximately 10mins for each subject |
* 16 subjects (the original version of the dataset included 19 subjects, but 3 were excluded from the group analysis for [[http://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingleOrig#Bad_subjects|various reasons]]) * 6 acquisition runs of approximately 10mins for each subject |
| Line 22: | Line 22: |
| * The subject has to judge the left-right symmetry of each stimulus * Total of nearly 300 trials in total for each of the 3 conditions |
* Participants had to judge the left-right symmetry of each stimulus * Total of nearly 300 trials for each of the 3 conditions |
| Line 31: | Line 31: |
| * MEG data have been "cleaned" using Signal-Space Separation as implemented in MaxFilter 2.1. * A Polhemus digitizer was used to digitise three fiducial points and a large number of other points across the scalp, which can be used to coregister the M/EEG data with the structural MRI image. |
* A Polhemus device was used to digitize three fiducial points and a large number of other points across the scalp, which can be used to coregister the M/EEG data with the structural MRI image. * Stimulation triggers: The triggers related with the visual presentation are saved in the STI101 channel, with the following event codes (bit 3 = face, bit 4 = unfamiliar, bit 5 = scrambled): * Famous faces: 5 (00101), 6 (00110), 7 (00111) * Unfamiliar faces: 13 (01101), 14 (01110), 15 (01111) * Scrambled images: 17 (10001), 18 (10010), 19 (10011) * Delays between the trigger in STI101 and the actual presentation of stimulus: '''34.5ms''' * MaxFilter/SSS: The data repository includes both the raw MEG recordings and the data cleaned using Signal-Space Separation as implemented in Elekta MaxFilter 2.2. The data was collected with continuous head localization and cannot be processed easily without running the algorithms SSS or tSSS. Brainstorm currently does not offer any free alternative to MaxFilter, therefore in this tutorial we will import the recordings already processed with MaxFilter's SSS, available in the "derivatives" folder of the BIDS architecture. To save disk space, will not download the raw MEG recordings. This means we will not get any of the additional files available in the BIDS structure (headshape, events, channels) but it doesn't matter because all the information is directly available in the .fif files. * Empty-room recordings: The data distribution includes MEG noise recordings acquired around the dates of the experiment, processed with MaxFilter 2.2 in the same way as the experimental data. The acquisition dates were removed from the subjects' recordings as part of the deidentification procedure, therefore it is not posssible to match by date the empty-room sessions with the subjects' sessions. The correspondance between subject name and closest empty-room session is available in the BIDS meta-data: * sub-emptyroom/ses-20090409: sub-01 * sub-emptyroom/ses-20090506: sub-02 * sub-emptyroom/ses-20090511: sub-03 * sub-emptyroom/ses-20090515: sub-05, sub-06, sub-07, sub-08, sub-09 * sub-emptyroom/ses-20090518: sub-04 * sub-emptyroom/ses-20090601: sub-11, sub-12, sub-13 * sub-emptyroom/ses-20091126: sub-14 * sub-emptyroom/ses-20091208: sub-15, sub-16 |
| Line 35: | Line 49: |
| * MRI data acquired on a 3T Siemens TIM Trio: 1x1x1mm T1-weighted structural MRI * Processed with FreeSurfer 5.3 |
* MRI data acquired on a 3T Siemens TIM Trio: 1x1x1mm T1-weighted structural MRI. * The face was removed from the structural images for anonymization purposes. * Processed with FreeSurfer 5.3. |
| Line 39: | Line 54: |
| * '''Requirements''': You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer. * The data is hosted on this FTP site (use an FTP client such as FileZilla, not your web browser): <<BR>>ftp://ftp.mrc-cbu.cam.ac.uk/personal/rik.henson/wakemandg_hensonrn/ * Download only the following folders (about 75Gb): * '''EmptyRoom''': Contains 3 sub-directories of empty-room recordings of 3-5mins acquired at roughly the same time of year (spring 2009) as the 16 subjects. The sub-directory names are Year (first 2 digits), Month (second 2 digits) and Day (third 2 digits). Inside each are 2 raw *.fif files: one for which basic SSS has been applied by maxfilter in a similar manner to the subject data above, and one (*-noSSS.fif) for which SSS has not been applied (though the data have been passed through maxfilter just to convert to float format). * '''Publications''': Reference publications related with this dataset. * '''SubXX/T1''': T1 MR scan and digitized positions (fiducials and headpoints). * '''SubXX/MEEG''': MEG and EEG recordings in FIF format and triggers definition files. * '''README.TXT''': License and dataset description. * The output of the FreeSurfer segmentation of the T1 structural images are not part of this package. You can either process them by yourself, or download the result of the segmentation on this website. Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page, and download the file: '''sample_group_anat.zip'''<<BR>>Unzip this file in the same folder where you downloaded all the datasets. * Do not put these downloaded files in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself. * Start Brainstorm (Matlab scripts or stand-alone version), see the [[Installation]] page. |
* First, make sure you have enough space on your hard drive, at least '''400Gb''': * Raw files: '''180Gb''' * Processed files: '''220Gb''' * The entire dataset is available from the OpenNeuro website as one single .zip file: <<BR>>https://openneuro.org/datasets/ds000117 <<BR>><<BR>> {{attachment:ds000117_download.gif}} * You will probably not manage to download 180Gb in one shot with your web browser: if it stops for any reason, you cannot resume the download. Even if you manage to download this gigantic .zip file, there is a chance the archive manager on your operating system cannot handle such a large file. * A safer alternative is to use the Amazon AWS command line tools: * Install AWS CLI: https://aws.amazon.com/cli/ (no need to create an account) * Linux/MacOS: * In a terminal, type: pip install awscli * Windows: * Download and install the program (64bit version, most likely) * Open a command line terminal (Windows menu, type "cmd", enter) * Go to the AWSCLI install folder (type: cd "C:\Program Files\Amazon\AWSCLI") * Start the download (synchronization with an Amazon S3 repository): * Create a new folder "ds000117" somewhere on a large hard drive (NOT in the Brainstorm program or database folders), referred to below as <target_directory>. * In the same terminal, type: * aws s3 sync --no-sign-request s3://openneuro.org/ds000117 <target_directory> * In this tutorial, we will use only the derivatives folder (85Gb), if you need to make some space on your hard drive you can delete all the other folders. Files we will use here: * FreeSurfer anatomy: /derivatives/freesurfer/sub-XX/ses-mri/anat/* * MEG recordings (SSS): /derivatives/meg_derivatives/sub-XX/ses-meg/meg/*.fif * Empty-room (SSS): /derivatives/meg_derivatives/sub-emptyroom/ses-meg/meg/*.fif * Start Brainstorm (Matlab scripts or stand-alone version). For help, see the [[Installation]] page. |
| Line 56: | Line 89: |
| This dataset is formatted following the [[https://www.biorxiv.org/content/early/2017/08/08/172684|BIDS-MEG specifications]], therefore we could import all the [[http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer|relevant information]] (MRI, FreeSurfer segmentation, MEG+EEG recordings) in just one click, with the menu '''File > Load protocol''' > '''Import BIDS dataset''', as described in the online tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/RestingOmega#BIDS_specifications|MEG resting state & OMEGA database]]. However, because your own data might not be organized following the BIDS standards, in this tutorial we preferred illustrating all the detailed steps for importing the data rather than the BIDS shortcut. Plus, we will need some additional steps that are not part of the standard import, because of the data anonymization (MRI were defaced and acquisition dates removed). This page explains how to import and process the first run of '''subject''' '''#01''' only. All the other files will have to be processed in the same way. |
|
| Line 57: | Line 94: |
| * Right-click on the TutorialAuditory folder > New subject > '''Sub01''' * Leave the default options you set for the protocol * Right-click on the subject node > Import anatomy folder: |
* Right-click on the TutorialVisual folder > New subject > '''sub-01''' * Leave the default options you defined for the protocol. * Right-click on the subject node > '''Import anatomy folder''': |
| Line 61: | Line 98: |
| * Select the folder: '''Anatomy/Sub01''' (from sample_group_anat.zip) | * Select the folder: '''derivatives/freesurfer/sub-01/ses-mri/anat''' |
| Line 63: | Line 100: |
| * In the coordinates panel, click on "'''Click here to compute MNI transformation'''". <<BR>>This step should take a few minutes, after it you should see the three referencial points AC, PC and IH. Additionally, you should have access to relatively accurate MNI coordinates. <<BR>><<BR>> {{attachment:anatomy_fiducials.gif||height="216",width="450"}} * To use the fiducials defined by the authors, open the file: '''Anatomy/Sub01/mri_fids.txt ''' * Click on the the button''' [Set the current coordinates]'''. * In the line "'''MNI coordinates'''", enter the position for the nasion ('''NAS''') from the fiducials file. * * Set the 6 required fiducial points (indicated in MRI coordinates): * NAS: x=127, y=213, z=139 * LPA: x=52, y=113, z=96 * RPA: x=202, y=113, z=91 * At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface. |
* When asked to select the anatomical fiducials, click on "'''Compute MNI transformation'''". This will register the MRI volume to an [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|MNI template]] with an affine transformation, using SPM functions embedded in Brainstorm (spm_maff8.m). This will also place default fiducial points NAS/LPA/RPA in the MRI, based on MNI coordinates. We will use digitized head points for the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Automatic_registration|MEG-MRI coregistration]], therefore we don't need accurate anatomical positions here. * Note that if you don't have a good digitized head shape for the subject, or if the final registration MEG-MRI doesn't look good with this head shape, you should not skip this step and mark accurate NAS/LPA/RPA fiducial points in the MRI, using the same anatomical convention that was used during the MRI acquisition. * Then click on [Save] to start the import. * At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). Otherwise, double-click on it to select it as the default cortex surface. Do not worry about the big holes in the head surface, parts of MRI have been remove voluntarily for anonymization purposes.<<BR>><<BR>> {{attachment:anatomy_import.gif||width="613",height="384"}} * All the anatomical atlases [[Tutorials/LabelFreeSurfer|generated by FreeSurfer]] were automatically imported: the cortical atlases (Desikan-Killiany, Mindboggle, Destrieux, Brodmann) and the sub-cortical regions (ASEG atlas). <<BR>><<BR>> {{attachment:anatomy_atlas.gif||width="550",height="211"}} == Access the recordings == === Link the recordings === We need to attach the continuous .fif files containing the recordings to the database. * Switch to the "functional data" view. * Right-click on the subject folder > '''Review raw file'''. * Select the file format: "'''MEG/EEG: Neuromag FIFF (*.fif)'''" * Go to the folder: '''derivatives/meg_derivatives/sub-01/ses-meg/meg''' * Select file: '''sub-01_ses-meg_task-facerecognition_run-01_proc-sss_meg.fif''' <<BR>><<BR>> {{attachment:review_raw.gif||width="583",height="205"}} * Events:''' Ignore'''. We will read the stimulus triggers later.<<BR>><<BR>> {{attachment:review_ignore.gif||width="330",height="186"}} * Refine registration now? '''NO''' <<BR>>The head points that are available in the FIF files contain all the points that were digitized during the MEG acquisition, including the ones corresponding to the parts of the face that have been removed from the MRI. If we run the fitting algorithm, all the points around the nose will not match any close points on the head surface, leading to a wrong result. We will first remove the face points and then run the registration manually. === Channel classification === A few non-EEG channels are mixed in with the EEG channels, we need to change this before applying any operation on the EEG channels. * Right-click on the channel file > '''Edit channel file'''. Double-click on a cell to edit it. * Change the type of '''EEG062''' to '''EOG '''(electrooculogram). * Change the type of '''EEG063 '''to '''ECG '''(electrocardiogram). * Change the type of '''EEG061''' and '''EEG064''' to '''NOSIG''' (or any other non-informative type). Close the window and save the modifications. <<BR>><<BR>> {{attachment:channel_edit.gif||width="561",height="252"}} === MRI registration === At this point, the registration MEG/MRI is based only on the three anatomical landmarks NAS/LPA/RPA, which are not even accurately set (we used default MNI positions). All the MRI scans were anonymized (defaced) so all the head points below the nasion cannot be used. We will try to refine this registration using the additional head points that were digitized above the nasion. * Right-click on the channel file > '''Digitized head points > Remove points below nasion'''. <<BR>>(42 points removed, 95 head points left)<<BR>><<BR>> {{attachment:channel_remove.gif||width="329",height="194"}} * Right-click on the channel file > '''MRI registration > Refine using head points'''.<<BR>><<BR>> {{attachment:channel_refine.gif||width="294",height="173"}} * MEG/MRI registration, before (left) and after (right) this automatic registration procedure: <<BR>><<BR>> {{attachment:registration.gif||width="236",height="209"}} {{attachment:registration_final.gif||width="237",height="208"}} * Right-click on the channel file > '''MRI registration > EEG: Edit'''.<<BR>>Click on ['''Project electrodes on surface'''], then close the figure to save the modifications.<<BR>><<BR>> {{attachment:channel_project.gif||width="477",height="207"}} === Read stimulus triggers === We need to read the stimulus markers from the STI channels. The following tasks can be done in an interactive way with menus in the Record tab, as in the introduction tutorials. We will illustrate here how to do this with the pipeline editor, it will be easier to batch it for all the runs and all the subjects. * In Process1, select the "Link to raw file", click on [Run]. * Select process '''Events > Read from channel''', Channel: '''STI101''', Detection mode: '''Bit'''.<<BR>>Do not execute the process, we will add other processes to classify the markers.<<BR>><<BR>> {{attachment:raw_read_events.gif||width="499",height="290"}} * We want to create three categories of events, based on their numerical codes: * '''Famous faces''': 5 (00101), 6 (00110), 7 (00111) => Bit 3 only * '''Unfamiliar faces''': 13 (01101), 14 (01110), 15 (01111) => Bit 3 and 4 * '''Scrambled images''': 17 (10001), 18 (10010), 19 (10011) => Bit 5 only * We will start by creating the category "Unfamiliar" (combination of events "3" and "4") and remove the remove the initial events. Then we just have to rename the remaining "3" in "Famous", and all the "5" in "Scrambled". * Add process '''Events > Group by name''': "'''Unfamiliar=3,4'''", Delay=0, '''Delete original events''' * Add process '''Events > Rename event''': 3 => Famous * Add process '''Events > Rename event''': 5 => Scrambled <<BR>><<BR>> {{attachment:events_merge.gif||width="532",height="402"}} * Add process '''Events > Add time offset''' to correct for the presentation delays:<<BR>>Event names: "'''Famous, Unfamiliar, Scrambled'''", Time offset = '''34.5ms'''<<BR>><<BR>> {{attachment:events_offset.gif||width="348",height="355"}} * Finally run the script. Double-click on the recordings to make sure the labels were detected correctly. You can delete the unwanted events that are left in the recordings (1,2,9,13). Edit the colors if you don't like the default ones.<<BR>><<BR>> {{attachment:events_display.gif||width="595",height="194"}} == Pre-processing == === Spectral evaluation === * Keep the "Link to raw file" in Process1. * Run process '''Frequency > Power spectrum density (Welch)''' with the options illustrated below. <<BR>><<BR>> {{attachment:psd_process.gif||width="669",height="314"}} * Right-click on the PSD file > Power spectrum. <<BR>><<BR>> {{attachment:psd_plot.gif||width="408",height="235"}} * The MEG sensors look awful, because of one small segment of data located around 248s. Open the MEG recordings and scroll to 248s (just before the first Unfamiliar event). <<BR>><<BR>> {{attachment:psd_error.gif||width="527",height="183"}} * In these recordings, the continuous head tracking was activated, but it starts only at the time of the stimulation (248s) while the acquisition of the data starts 22s before (226s). The first 22s do not have head localization coils (HPI) coils activity and are not corrected by MaxFilter. After 248s, the HPI coils are on, and MaxFilter filters them out. The transition between the two states is not smooth and creates important distortions in the spectral domain. For a proper evaluation of the recordings, we should '''compute the PSD only after the HPI coils are turned on'''. * Run process '''Events > Detect cHPI activity (Elekta)'''. This detects the changes in the cHPI activity from channel STI201 and marks all the data without head localization as bad. <<BR>><<BR>> {{attachment:psd_badsegment.gif||width="503",height="234"}} * Re-run the process '''Frequency > Power spectrum density (Welch)'''. All the bad segments are excluded from the computation, therefore the PSD is now estimated only with the data '''after 248s'''. <<BR>><<BR>> {{attachment:psd_fix.gif||width="668",height="283"}} * Observations: * Three groups of sensors, from top to bottom: EEG, MEG gradiometers, MEG magnetometers. * Power lines: '''50 '''Hz and harmonics * Alpha peak around 10 Hz * Artifacts due to Elekta electronics (HPI coils): '''293'''Hz, '''307'''Hz, '''314'''Hz, '''321'''Hz, '''328'''Hz. * Peak from unknown source at '''103.4Hz''' in the MEG only. * Suspected bad EEG channels: '''EEG016''' * Close all the windows. === Remove line noise === * Keep the "Link to raw file" in Process1. * Select process '''Pre-process > Notch filter''' to remove the line noise (50-200Hz).<<BR>>Add the process '''Frequency > Power spectrum density (Welch)'''. <<BR>><<BR>> {{attachment:notch_process.gif||width="562",height="262"}} * Double-click on the PSD for the new continuous file to evaluate the quality of the correction. <<BR>><<BR>> {{attachment:notch_result.gif||width="566",height="215"}} * Close all the windows (use the [X] button at the top-right corner of the Brainstorm window). === EEG reference and bad channels === * Right-click on link to the processed file ("Raw | notch(50Hz ...") > '''EEG > Display time series'''. * Select channel '''EEG016''' and mark it as '''bad''' (using the popup menu or pressing the Delete key). <<BR>><<BR>> {{attachment:channel_bad.gif||width="587",height="214"}} * In the Record tab, menu '''Artifacts > Re-reference EEG''' > "AVERAGE". <<BR>><<BR>> {{attachment:channel_ref.gif||width="535",height="270"}} * At the end, the window "select active projectors" is open to show the new re-referencing projector. Just close this window. To get it back, use the menu Artifacts > Select active projectors. == Artifact detection == === Heartbeats: Detection === * Empty the Process1 list (right-click > Clear list). * Drag and drop the continuous processed file ("Raw | notch(50Hz...)") to the Process1 list. * Run process''' Events > Detect heartbeats''': Channel name='''EEG063''', All file, Event name=cardiac <<BR>><<BR>> {{attachment:detect_cardiac.gif||width="310",height="230"}} === Eye blinks: Detection === * In many of the other tutorials, we detect the blinks and remove them with SSP. In this experiment, we are particularly interested in the subject's response to seeing the stimulus. Therefore we will exclude from the analysis all the recordings contaminated with blinks or other eye movements. * Run process '''Events > Detect events above threshold''': <<BR>>Event name='''blink_BAD''', Channel='''EEG062''', All file, Maximum threshold='''100''', Threshold units='''uV''', Filter='''[0.30,20.00]Hz''', '''Use absolute value of signal'''.<<BR>><<BR>> {{attachment:detect_blinks.gif||width="285",height="455"}} * In other tutorials, we used the process "Detect eye blinks" which creates simple events indicating the peak of the EOG signal during a blink. In this study, we preferred using this process "Detect events above threshold" because it creates extended events marking as bad all the segment for which the EOG value is above a given threshold. It's more manual but it ensures we really exclude all the ocular activity. * Inspect visually the two new cateogries of events: cardiac and blink_BAD.<<BR>><<BR>> {{attachment:detect_display.gif||width="655",height="237"}} * Close all the windows (using the [X] button). === Heartbeats: Correction with SSP === * Keep the "Raw | notch" file selected in Process1. * Select process '''Artifacts > SSP: Heartbeats''' > Sensor type: '''MEG MAG''' * Add process '''Artifacts > SSP: Heartbeats''' > Sensor type: '''MEG GRAD''' - Run the execution <<BR>><<BR>> {{attachment:ssp_ecg_process.gif||width="293",height="225"}} * Double-click on the continuous file to show all the MEG sensors. <<BR>>In the Record tab, select sensors "Left-temporal". * Menu Artifacts > Select active projectors. * In category '''cardiac/MEG MAG''': Select '''component #1''' and view topography. * In category '''cardiac/MEG GRAD''': Select '''component #1 '''and view topography. <<BR>><<BR>> {{attachment:ssp_ecg_topo.gif}} * Make sure that selecting the two components removes the cardiac artifact. Then click '''[Save]'''. === Additional bad segments === * Process1>Run: Select the process "'''Events > Detect other artifacts'''". This should be done separately for MEG and EEG to avoid confusion about which sensors are involved in the artifact. <<BR>><<BR>> {{attachment:detect_other.gif||width="486",height="253"}} * Display the MEG sensors. Review the segments that were tagged as artifact, determine if each event represents an artifact and then mark the time of the artifact as BAD. This can be done by selecting the time window around the artifact, then right-click > '''Reject time segment'''. Note that this detection process marks 1-second segments but the artifact can be shorter.<<BR>><<BR>> {{attachment:artifacts_mark_bad.png||width="640"}} * Once all the events in the two categories are reviewed and bad segments are marked, the two categories (1-7Hz and 40-240Hz) can be deleted. If want to mark as bad most of the events in a category, another solution is to add the tag BAD to its name (or menu Events > Mark group as bad), and remove from it the segments you don't want to reject. * Do this detection and review again for the EEG. === SQUID jumps === MEG signals recorded with Elekta-Neuromag systems frequently contain SQUID jumps ([[http://neuroimage.usc.edu/brainstorm/Tutorials/BadSegments?highlight=(squid)#Elekta-Neuromag_SQUID_jumps|more information]]). These sharp steps followed by a change of baseline value are easy to identify visually but more complicated to detect automatically. The process "Detect other artifacts" usually detects most of them in the category "1-7Hz". If you observe that some are skipped, you can try re-running it with a higher sensitivity. It is important to review all the sensors and all the time in each run to be sure these events are marked as bad segments. . {{attachment:detect_jumps.gif||width="547",height="214"}} == Epoching and averaging == === Import epochs === * Keep the "Raw | notch" file selected in Process1. * Select process: '''Import > Import recordings > Import MEG/EEG: Events''' (do not run now)<<BR>>Event names "Famous, Unfamiliar, Scrambled", All file, Epoch time=[-500,1200]ms * Add process: '''Pre-process > Remove DC offset''': Baseline=[-500,-0.9]ms - Run execution.<<BR>><<BR>> {{attachment:import_epochs.gif||width="647",height="399"}} === Average by run === * In Process1, select all the imported trials. * Run process: '''Average > Average files''': '''By trial groups (folder average)''' <<BR>><<BR>> {{attachment:average_process.gif||width="478",height="504"}} === Review EEG ERP === * EEG evoked response (famous, scrambled, unfamiliar), display with low-pass filter at 32Hz: <<BR>><<BR>> {{attachment:average_topo.gif||width="684",height="392"}} * Open the [[Tutorials/ChannelClusters|Cluster tab]] and create a cluster with the channel '''EEG065''' (button [NEW IND]).<<BR>><<BR>> {{attachment:cluster_create.gif||width="569",height="166"}} * Select the cluster, select the three average files, right-click > '''Clusters time series''' (Overlay:Files). <<BR>><<BR>> {{attachment:cluster_display.gif||width="658",height="220"}} * Basic observations for EEG065 (right parieto-occipital electrode): * Around 170ms (N170): greater negative deflection for Famous than Scrambled faces. * After 250ms: difference between Famous and Unfamiliar faces. == Source estimation == === MEG noise covariance: Empty room recordings === The minimum norm model we will use next to estimate the source activity can be improved by modeling the the noise contaminating the data. The introduction tutorials explain how to [[Tutorials/NoiseCovariance|estimate the noise covariance]] in different ways for EEG and MEG. For the MEG recordings we will use the empty room measurements we have, and for the EEG we will compute it from the pre-stimulus baselines we have in all the imported epochs. There are 8 empty room files available in this dataset. For each subject, we will use only one file, the one that was acquired at the closest date. We will now import and process all the empty room recordings simultaneously, even if only one is needed by the current subject. Later, for each subject we will select the most appropriate noise covariance matrix. * Create a new subject: '''sub-emptyroom''' * Right-click on the new subject > '''Review raw file'''.<<BR>>Select all the folders in: '''derivatives/meg_derivatives/sub-emptyroom '''(processed with MaxFilter/SSS)<<BR>>Do not apply default transformation, Ignore event channel.<<BR>><<BR>> {{attachment:noise_review.gif||width="516",height="180"}} * Select all these new files in Process1 and run process '''Pre-process > Notch filter''': '''50 100 150 200Hz'''. When using empty room measurements to compute the noise covariance, they must be processed exactly in the same way as the other recordings (same MaxFilter parameters, same frequency filters). <<BR>><<BR>> {{attachment:noise_notch.gif||width="634",height="242"}} * Delete all the original noise files and keep only the filtered ones. * You need now to identify the noise file that was recorded the closest to the subject's recordings. In general, you can read the acquisition date of a file in the tooptip that is displayed when you leave you mouse for a while over its folder. However, the subjects' .fif files in this dataset were deidentified, we do not have access to the real acquisition dates. <<BR>><<BR>> {{attachment:noise_closest.gif}} * From the BIDS meta-data, we know that the appropriate empty-room for sub-01 is ses-20090409, from April 9th 2009. We will set manually the acquisition date of all the runs in sub-01 to this same date, so that we can use the automatic matching by date in the scripts. * Right-click on the folder containing all the imported epochs > File > '''Set acquisition date'''. Enter 09-04-2009 (April 9th, 2009). <<BR>><<BR>> {{attachment:noise_setdate.gif||width="568",height="317"}} * Right-click on the noise recordings 2009-04-09 > '''Noise covariance > Compute from recordings''': <<BR>><<BR>> {{attachment:noise_compute.gif||width="641",height="276"}} * Finally copy the noise covariance from sub-emptyroom to the folder with imported epochs in sub-01. Use the popup menus File>Copy/Paste, or the keyboard shortcuts Ctrl+C/Ctrl+V.<<BR>><<BR>> {{attachment:noise_copy.gif||width="685",height="246"}} === EEG noise covariance: Pre-stimulus baseline === * In folder sub-01/run-01_notch, select all the imported the imported trials, right-click > '''Noise covariance > Compute from recordings''', Time=[-500,-0.9]ms, '''EEG only''', '''Merge'''. {{attachment:noise_eeg.gif||width="650",height="280"}} * This computes the noise covariance only for EEG, and combines it with the existing MEG information. {{attachment:noise_display.gif||width="375",height="223"}} * To generate the corresponding analysis script, we will use a different strategy. We will first import all the subjects, compute the EEG noise covariance for all the runs, then compute the MEG noise covariance matrix for all the empty room files at once with the process "Sources > Compute covariance" and finally merge the MEG+EEG noise covariances. When the acquisition dates are correctly set, we can use the options "Copy to other folders/subjects" and "Match noise and subject recordings by acquisition date". <<BR>><<BR>> {{attachment:noise_automatch.gif||width="491",height="427"}} === BEM layers === We will compute a [[Tutorials/TutBem|BEM forward model]] to estimate the brain sources from the EEG recordings. For this, we need some layers defining the separation between the different tissues of the head (scalp, inner skull, outer skull). * Go to the anatomy view (first button above the database explorer). * Right-click on the subject folder > '''Generate BEM surfaces''': The number of vertices to use for each layer depends on your computing power and the accuracy you expect. You can try for instance with '''1082 vertices '''(scalp) and '''642 vertices '''(outer skull and inner skull). <<BR>><<BR>> {{attachment:anatomy_bem.gif||width="603",height="232"}} === Forward model: EEG and MEG === * Go back to the functional view (second button above the database explorer). * Model used: Overlapping spheres for MEG, OpenMEEG BEM for EEG ([[Tutorials/HeadModel|more information]]). * In folder sub-01/run_01_notch, right-click on the channel file > '''Compute head model'''.<<BR>>Keep all the default options . Expect this to take a while...<<BR>><<BR>> {{attachment:headmodel_compute.gif||width="643",height="313"}} === Inverse model: Minimum norm estimates === * Right-click on head model > '''Compute sources [2018]''': '''MEG MAG + GRAD''' (default options) <<BR>><<BR>> {{attachment:sources_compute.gif||width="626",height="406"}} * Right-click on head model > '''Compute sources [2018]''': '''EEG''' (default bad channels). * At the end we have two inverse operators, that are shared for all the files of the run (single trials and averages). If we wanted to look at the run-level source averages, we could normalize the source maps with a Z-score wrt baseline. In this tutorial, we will first average across runs and normalize the subject-level averages. This will be done in the next tutorial (group analysis).<<BR>><<BR>> {{attachment:sources_files.gif||width="243",height="147"}} == Time-frequency analysis == We will compute the time-frequency decomposition of each trial using Morlet wavelets, and average the power of the Morlet coefficients for each condition and each run separately. We will restrict the computation to the MEG magnetometers and the EEG channels to limit the computation time and disk usage. * In Process1, select the imported trials "Famous" for run#01. * Run process '''Frequency > Time-frequency (Morlet wavelets)''': Sensor types=MEG MAG,EEG <<BR>>Not normalized, Frequency='''Log(6:20:60)''', Measure=Power, '''Save average''' <<BR>><<BR>> {{attachment:tf_process.gif||width="628",height="479"}} * Double-click on the file to display it. In the Display tab, select the option "Hide edge effects" to exclude from the display all the values that could not be estimated in a reliable way. Let's extract only the good values from this file (-200ms to +900ms). <<BR>><<BR>> {{attachment:tf_display.gif||width="270",height="155"}} * In Process1, select the time-frequency file. * Run process '''Extract > Extract time''': Time window='''[-200, 900]ms''', '''Overwrite input files''' <<BR>><<BR>> {{attachment:tf_cut.gif||width="411",height="197"}} * Display the file again, observe that all the possibly bad values are gone. <<BR>><<BR>> {{attachment:tf_cutdisplay.gif||width="410",height="178"}} * You can display all the sensors at once (MEG MAG or EEG): right-click > 2D Layout (maps).<<BR>><<BR>> {{attachment:tf_2dlayout.gif||width="653",height="270"}} * Repeat these steps for other conditions (Scrambled and Unfamiliar) and the other runs (2-6). There is no way with this process to compute all the averages at once, as it is with the process "Average files". This will be easier to run from a script. * If we wanted to look at the run-level source averages, we could normalize these time-frequency maps. In this tutorial, we will first average across runs and normalize the subject-level averages. This will be done in the next tutorial (group analysis). == Scripting == We have now all the files we need for the group analysis ([[Tutorials/VisualGroup|next tutorial]]). We need to repeat the same operations for all the runs and all the subjects. Some of these steps are '''fully automatic''' and take a lot of time (filtering, computing the forward model), they should be executed from a script. However, we recommend you always '''review manually''' some of the pre-processing steps (selection of the bad segments and bad channels, SSP/ICA components). Do not trust blindly any fully automated cleaning procedure. For the strict reproducibility of this analysis, we provide a script that processes all the 16 subjects: '''brainstorm3/toolbox/script/tutorial_visual_single.m''' (execution time: 10-30 hours)<<BR>>Report for the first subject: [[http://neuroimage.usc.edu/bst/examples/report_TutorialVisual_sub-01.html|report_TutorialVisual_sub-01.html]] <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:500px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_visual_single.m")>><<HTML(</div >)>> <<BR>>You should note that this is not the result of a fully automated procedure. The bad channels were identified manually and are defined for each run in the script. The bad segments were detected automatically, confirmed manually for each run and saved in external files, then exported as text ans copied at the end of this script. All the process calls (bst_process) were generated automatically using with the script generator (menu '''Generate .m script''' in the pipeline editor). Everything else was added manually (loops, bad channels, file copies). <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=&next=Tutorials/VisualGroup")>> |
MEG visual tutorial: Single subject (BIDS)
Authors: Francois Tadel, Elizabeth Bock.
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "Multimodal, Multisubject data fusion". We use here a recent update of this dataset, reformatted to follow the Brain Imaging Data Structure (BIDS), a standard for neuroimaging data organization. It is part of a collective effort to document and standardize MEG/EEG group analysis, see Frontier's research topic: From raw MEG/EEG to publication: how to perform MEG/EEG group analysis with free academic software.
The data processed here consists of simultaneous MEG/EEG recordings from 16 participants performing a simple visual recognition task from presentations of famous, unfamiliar and scrambled faces. The analysis is split in two tutorial pages: the present tutorial describes the detailed interactive analysis of one single subject; the second tutorial describes batch processing and group analysis of all 16 participants.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Contents
License
This dataset was obtained from the OpenNeuro project (http://openneuro.org), accession #ds117. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data: Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset, Scientific Data (2015)
Any questions regarding the data, please contact: rik.henson@mrc-cbu.cam.ac.uk
Presentation of the experiment
Experiment
16 subjects (the original version of the dataset included 19 subjects, but 3 were excluded from the group analysis for various reasons)
- 6 acquisition runs of approximately 10mins for each subject
- Presentation of series of images: familiar faces, unfamiliar faces, phase-scrambled faces
- Participants had to judge the left-right symmetry of each stimulus
- Total of nearly 300 trials for each of the 3 conditions
MEG acquisition
Acquisition at 1100Hz with an Elekta-Neuromag VectorView system (simultaneous MEG+EEG).
- Recorded channels (404):
- 102 magnetometers
- 204 planar gradiometers
- 70 EEG electrodes recorded with a nose reference.
- A Polhemus device was used to digitize three fiducial points and a large number of other points across the scalp, which can be used to coregister the M/EEG data with the structural MRI image.
- Stimulation triggers: The triggers related with the visual presentation are saved in the STI101 channel, with the following event codes (bit 3 = face, bit 4 = unfamiliar, bit 5 = scrambled):
- Famous faces: 5 (00101), 6 (00110), 7 (00111)
- Unfamiliar faces: 13 (01101), 14 (01110), 15 (01111)
- Scrambled images: 17 (10001), 18 (10010), 19 (10011)
Delays between the trigger in STI101 and the actual presentation of stimulus: 34.5ms
MaxFilter/SSS: The data repository includes both the raw MEG recordings and the data cleaned using Signal-Space Separation as implemented in Elekta MaxFilter 2.2. The data was collected with continuous head localization and cannot be processed easily without running the algorithms SSS or tSSS. Brainstorm currently does not offer any free alternative to MaxFilter, therefore in this tutorial we will import the recordings already processed with MaxFilter's SSS, available in the "derivatives" folder of the BIDS architecture. To save disk space, will not download the raw MEG recordings. This means we will not get any of the additional files available in the BIDS structure (headshape, events, channels) but it doesn't matter because all the information is directly available in the .fif files.
Empty-room recordings: The data distribution includes MEG noise recordings acquired around the dates of the experiment, processed with MaxFilter 2.2 in the same way as the experimental data. The acquisition dates were removed from the subjects' recordings as part of the deidentification procedure, therefore it is not posssible to match by date the empty-room sessions with the subjects' sessions. The correspondance between subject name and closest empty-room session is available in the BIDS meta-data:
- sub-emptyroom/ses-20090409: sub-01
- sub-emptyroom/ses-20090506: sub-02
- sub-emptyroom/ses-20090511: sub-03
- sub-emptyroom/ses-20090515: sub-05, sub-06, sub-07, sub-08, sub-09
- sub-emptyroom/ses-20090518: sub-04
- sub-emptyroom/ses-20090601: sub-11, sub-12, sub-13
- sub-emptyroom/ses-20091126: sub-14
- sub-emptyroom/ses-20091208: sub-15, sub-16
Subject anatomy
- MRI data acquired on a 3T Siemens TIM Trio: 1x1x1mm T1-weighted structural MRI.
- The face was removed from the structural images for anonymization purposes.
Processed with FreeSurfer 5.3.
Download and installation
First, make sure you have enough space on your hard drive, at least 400Gb:
Raw files: 180Gb
Processed files: 220Gb
The entire dataset is available from the OpenNeuro website as one single .zip file:
https://openneuro.org/datasets/ds000117
- You will probably not manage to download 180Gb in one shot with your web browser: if it stops for any reason, you cannot resume the download. Even if you manage to download this gigantic .zip file, there is a chance the archive manager on your operating system cannot handle such a large file.
- A safer alternative is to use the Amazon AWS command line tools:
Install AWS CLI: https://aws.amazon.com/cli/ (no need to create an account)
- Linux/MacOS:
- In a terminal, type: pip install awscli
- Windows:
- Download and install the program (64bit version, most likely)
- Open a command line terminal (Windows menu, type "cmd", enter)
- Go to the AWSCLI install folder (type: cd "C:\Program Files\Amazon\AWSCLI")
- Linux/MacOS:
- Start the download (synchronization with an Amazon S3 repository):
Create a new folder "ds000117" somewhere on a large hard drive (NOT in the Brainstorm program or database folders), referred to below as <target_directory>.
- In the same terminal, type:
aws s3 sync --no-sign-request s3://openneuro.org/ds000117 <target_directory>
- In this tutorial, we will use only the derivatives folder (85Gb), if you need to make some space on your hard drive you can delete all the other folders. Files we will use here:
FreeSurfer anatomy: /derivatives/freesurfer/sub-XX/ses-mri/anat/*
- MEG recordings (SSS): /derivatives/meg_derivatives/sub-XX/ses-meg/meg/*.fif
- Empty-room (SSS): /derivatives/meg_derivatives/sub-emptyroom/ses-meg/meg/*.fif
Start Brainstorm (Matlab scripts or stand-alone version). For help, see the Installation page.
Select the menu File > Create new protocol. Name it "TutorialVisual" and select the options:
"No, use individual anatomy",
"No, use one channel file per condition".
Import the anatomy
This dataset is formatted following the BIDS-MEG specifications, therefore we could import all the relevant information (MRI, FreeSurfer segmentation, MEG+EEG recordings) in just one click, with the menu File > Load protocol > Import BIDS dataset, as described in the online tutorial MEG resting state & OMEGA database. However, because your own data might not be organized following the BIDS standards, in this tutorial we preferred illustrating all the detailed steps for importing the data rather than the BIDS shortcut. Plus, we will need some additional steps that are not part of the standard import, because of the data anonymization (MRI were defaced and acquisition dates removed).
This page explains how to import and process the first run of subject #01 only. All the other files will have to be processed in the same way.
- Switch to the "anatomy" view.
Right-click on the TutorialVisual folder > New subject > sub-01
- Leave the default options you defined for the protocol.
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: derivatives/freesurfer/sub-01/ses-mri/anat
- Number of vertices of the cortex surface: 15000 (default value)
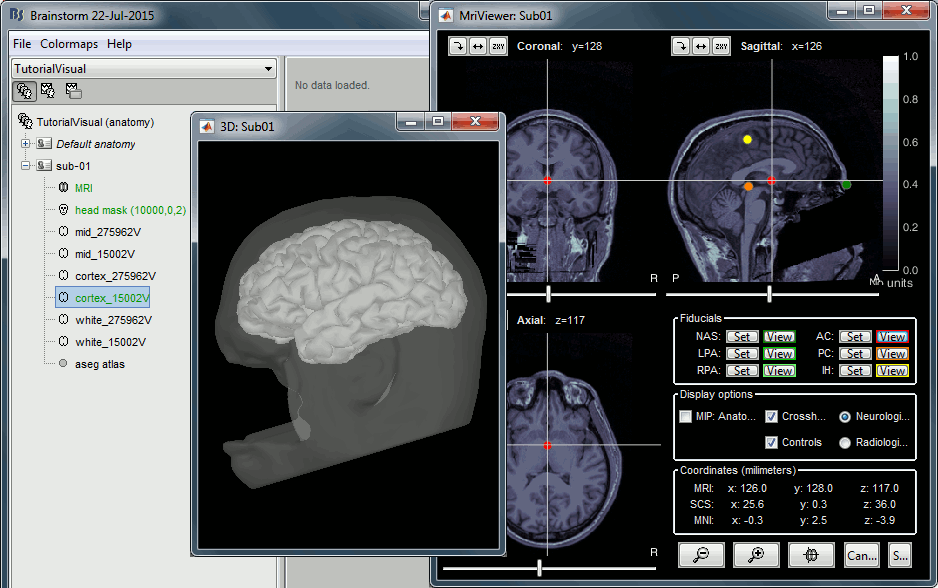
When asked to select the anatomical fiducials, click on "Compute MNI transformation". This will register the MRI volume to an MNI template with an affine transformation, using SPM functions embedded in Brainstorm (spm_maff8.m). This will also place default fiducial points NAS/LPA/RPA in the MRI, based on MNI coordinates. We will use digitized head points for the MEG-MRI coregistration, therefore we don't need accurate anatomical positions here.
- Note that if you don't have a good digitized head shape for the subject, or if the final registration MEG-MRI doesn't look good with this head shape, you should not skip this step and mark accurate NAS/LPA/RPA fiducial points in the MRI, using the same anatomical convention that was used during the MRI acquisition.
- Then click on [Save] to start the import.
At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). Otherwise, double-click on it to select it as the default cortex surface. Do not worry about the big holes in the head surface, parts of MRI have been remove voluntarily for anonymization purposes.
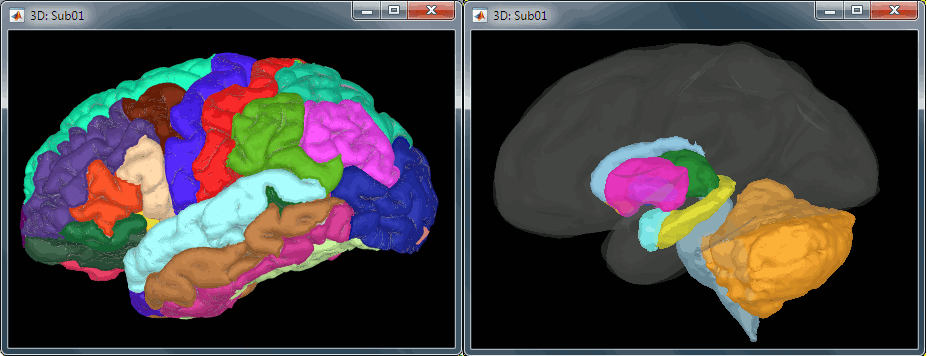
All the anatomical atlases generated by FreeSurfer were automatically imported: the cortical atlases (Desikan-Killiany, Mindboggle, Destrieux, Brodmann) and the sub-cortical regions (ASEG atlas).
Access the recordings
Link the recordings
We need to attach the continuous .fif files containing the recordings to the database.
- Switch to the "functional data" view.
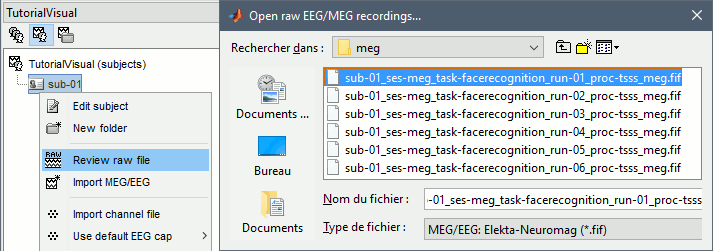
Right-click on the subject folder > Review raw file.
Select the file format: "MEG/EEG: Neuromag FIFF (*.fif)"
Go to the folder: derivatives/meg_derivatives/sub-01/ses-meg/meg
Select file: sub-01_ses-meg_task-facerecognition_run-01_proc-sss_meg.fif
Events: Ignore. We will read the stimulus triggers later.
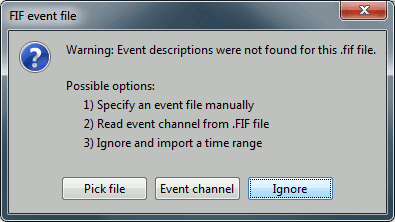
Refine registration now? NO
The head points that are available in the FIF files contain all the points that were digitized during the MEG acquisition, including the ones corresponding to the parts of the face that have been removed from the MRI. If we run the fitting algorithm, all the points around the nose will not match any close points on the head surface, leading to a wrong result. We will first remove the face points and then run the registration manually.
Channel classification
A few non-EEG channels are mixed in with the EEG channels, we need to change this before applying any operation on the EEG channels.
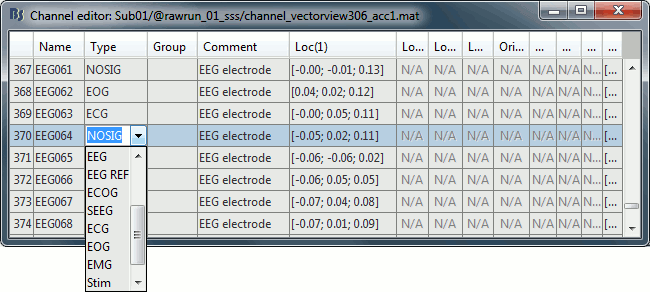
Right-click on the channel file > Edit channel file. Double-click on a cell to edit it.
Change the type of EEG062 to EOG (electrooculogram).
Change the type of EEG063 to ECG (electrocardiogram).
Change the type of EEG061 and EEG064 to NOSIG (or any other non-informative type). Close the window and save the modifications.
MRI registration
At this point, the registration MEG/MRI is based only on the three anatomical landmarks NAS/LPA/RPA, which are not even accurately set (we used default MNI positions). All the MRI scans were anonymized (defaced) so all the head points below the nasion cannot be used. We will try to refine this registration using the additional head points that were digitized above the nasion.
Right-click on the channel file > Digitized head points > Remove points below nasion.
(42 points removed, 95 head points left)
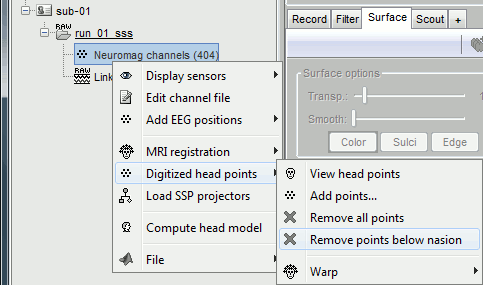
Right-click on the channel file > MRI registration > Refine using head points.
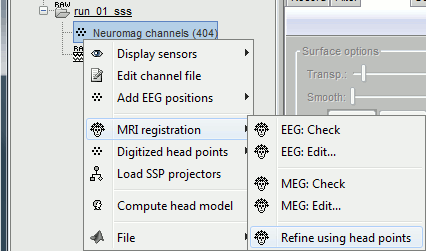
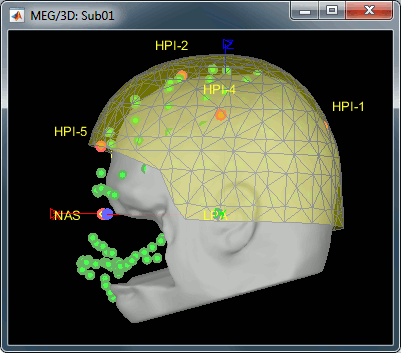
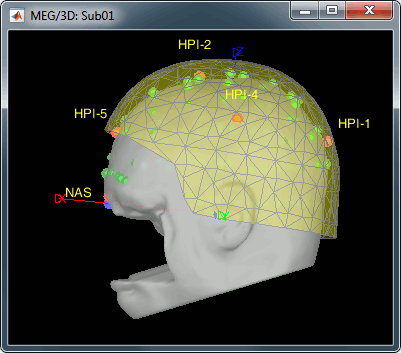
MEG/MRI registration, before (left) and after (right) this automatic registration procedure:
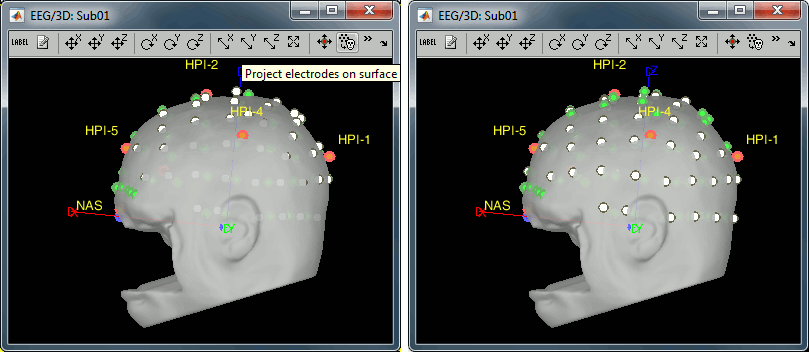
Right-click on the channel file > MRI registration > EEG: Edit.
Click on [Project electrodes on surface], then close the figure to save the modifications.
Read stimulus triggers
We need to read the stimulus markers from the STI channels. The following tasks can be done in an interactive way with menus in the Record tab, as in the introduction tutorials. We will illustrate here how to do this with the pipeline editor, it will be easier to batch it for all the runs and all the subjects.
- In Process1, select the "Link to raw file", click on [Run].
Select process Events > Read from channel, Channel: STI101, Detection mode: Bit.
Do not execute the process, we will add other processes to classify the markers.
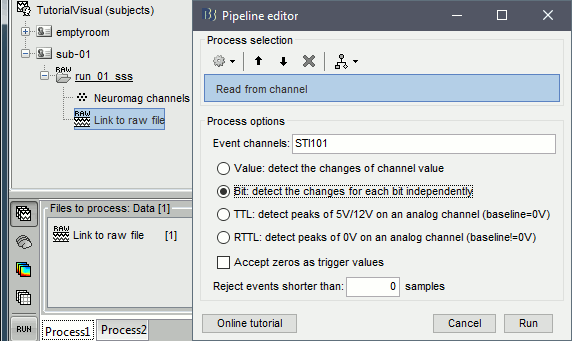
- We want to create three categories of events, based on their numerical codes:
Famous faces: 5 (00101), 6 (00110), 7 (00111) => Bit 3 only
Unfamiliar faces: 13 (01101), 14 (01110), 15 (01111) => Bit 3 and 4
Scrambled images: 17 (10001), 18 (10010), 19 (10011) => Bit 5 only
- We will start by creating the category "Unfamiliar" (combination of events "3" and "4") and remove the remove the initial events. Then we just have to rename the remaining "3" in "Famous", and all the "5" in "Scrambled".
Add process Events > Group by name: "Unfamiliar=3,4", Delay=0, Delete original events
Add process Events > Rename event: 3 => Famous
Add process Events > Rename event: 5 => Scrambled
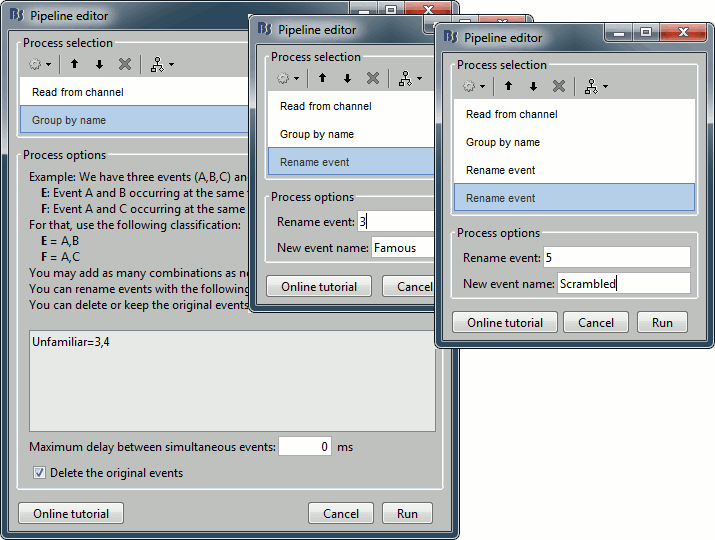
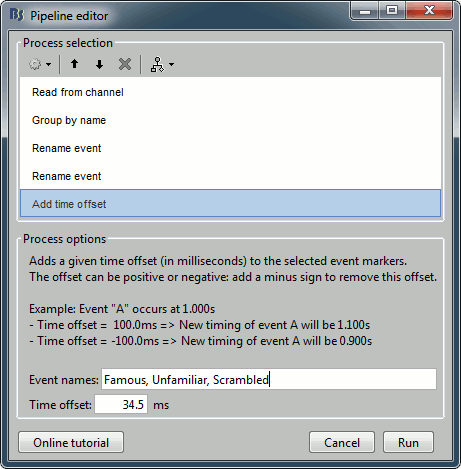
Add process Events > Add time offset to correct for the presentation delays:
Event names: "Famous, Unfamiliar, Scrambled", Time offset = 34.5ms
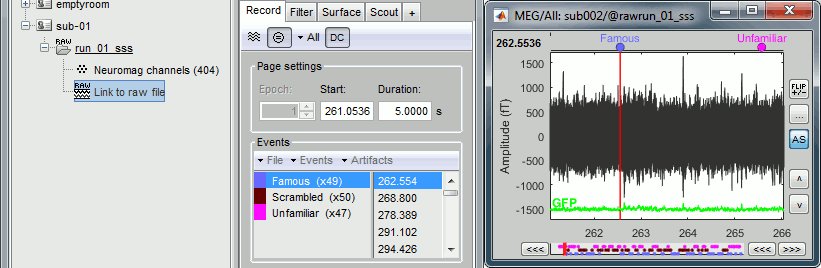
Finally run the script. Double-click on the recordings to make sure the labels were detected correctly. You can delete the unwanted events that are left in the recordings (1,2,9,13). Edit the colors if you don't like the default ones.
Pre-processing
Spectral evaluation
- Keep the "Link to raw file" in Process1.
Run process Frequency > Power spectrum density (Welch) with the options illustrated below.
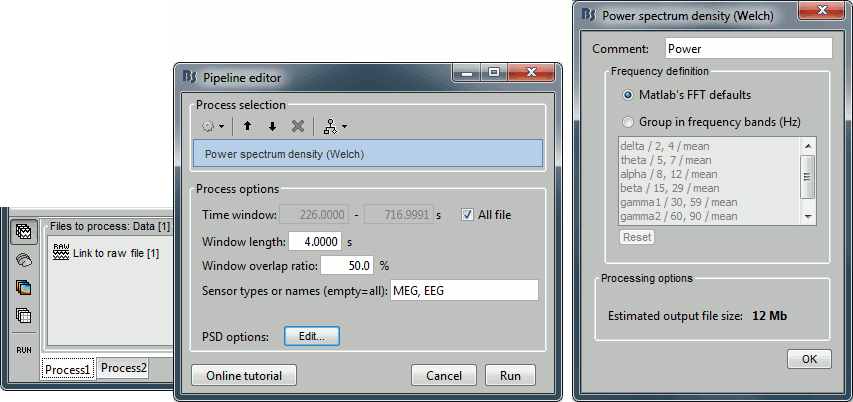
Right-click on the PSD file > Power spectrum.
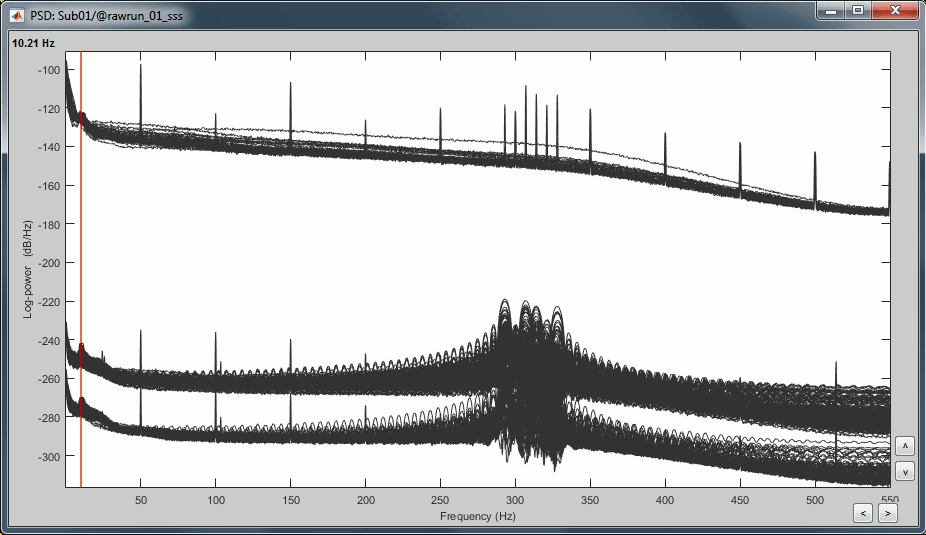
The MEG sensors look awful, because of one small segment of data located around 248s. Open the MEG recordings and scroll to 248s (just before the first Unfamiliar event).
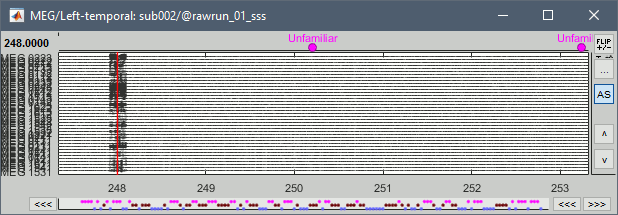
In these recordings, the continuous head tracking was activated, but it starts only at the time of the stimulation (248s) while the acquisition of the data starts 22s before (226s). The first 22s do not have head localization coils (HPI) coils activity and are not corrected by MaxFilter. After 248s, the HPI coils are on, and MaxFilter filters them out. The transition between the two states is not smooth and creates important distortions in the spectral domain. For a proper evaluation of the recordings, we should compute the PSD only after the HPI coils are turned on.
Run process Events > Detect cHPI activity (Elekta). This detects the changes in the cHPI activity from channel STI201 and marks all the data without head localization as bad.
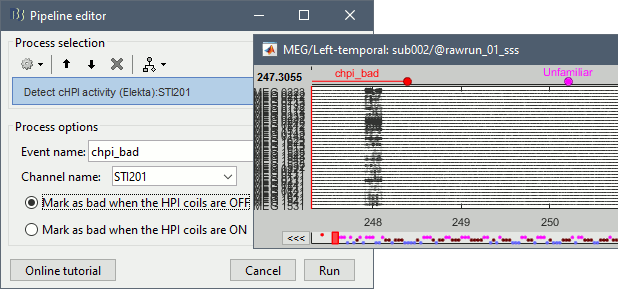
Re-run the process Frequency > Power spectrum density (Welch). All the bad segments are excluded from the computation, therefore the PSD is now estimated only with the data after 248s.
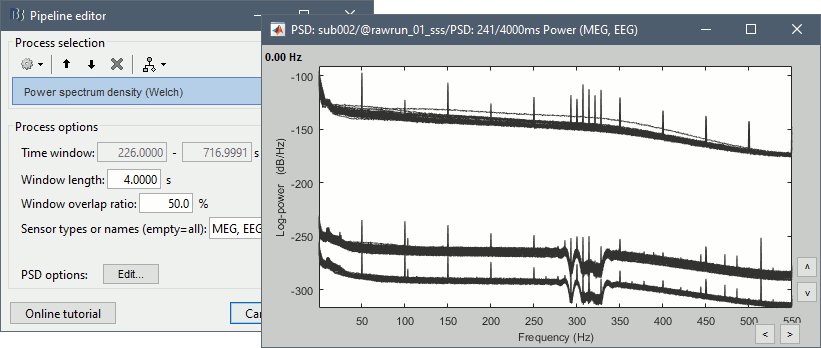
- Observations:
- Three groups of sensors, from top to bottom: EEG, MEG gradiometers, MEG magnetometers.
Power lines: 50 Hz and harmonics
- Alpha peak around 10 Hz
Artifacts due to Elekta electronics (HPI coils): 293Hz, 307Hz, 314Hz, 321Hz, 328Hz.
Peak from unknown source at 103.4Hz in the MEG only.
Suspected bad EEG channels: EEG016
- Close all the windows.
Remove line noise
- Keep the "Link to raw file" in Process1.
Select process Pre-process > Notch filter to remove the line noise (50-200Hz).
Add the process Frequency > Power spectrum density (Welch).
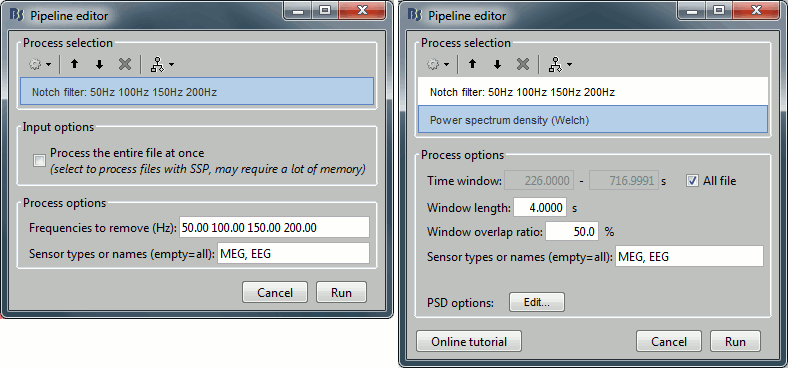
Double-click on the PSD for the new continuous file to evaluate the quality of the correction.
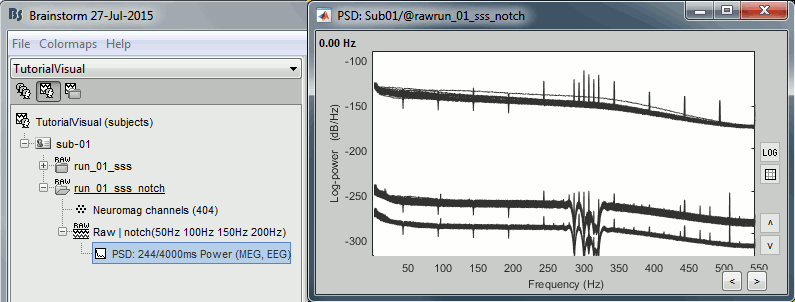
- Close all the windows (use the [X] button at the top-right corner of the Brainstorm window).
EEG reference and bad channels
Right-click on link to the processed file ("Raw | notch(50Hz ...") > EEG > Display time series.
Select channel EEG016 and mark it as bad (using the popup menu or pressing the Delete key).
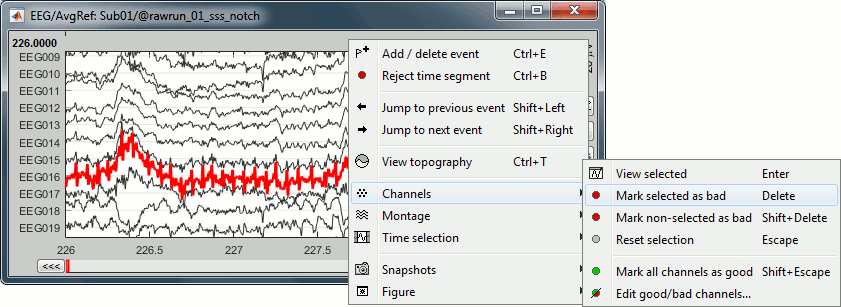
In the Record tab, menu Artifacts > Re-reference EEG > "AVERAGE".
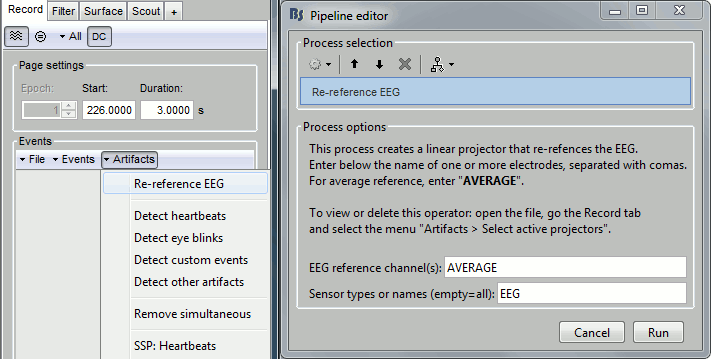
At the end, the window "select active projectors" is open to show the new re-referencing projector. Just close this window. To get it back, use the menu Artifacts > Select active projectors.
Artifact detection
Heartbeats: Detection
Empty the Process1 list (right-click > Clear list).
- Drag and drop the continuous processed file ("Raw | notch(50Hz...)") to the Process1 list.
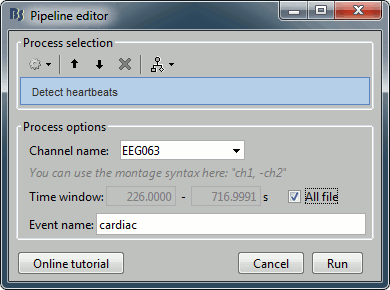
Run process Events > Detect heartbeats: Channel name=EEG063, All file, Event name=cardiac
Eye blinks: Detection
- In many of the other tutorials, we detect the blinks and remove them with SSP. In this experiment, we are particularly interested in the subject's response to seeing the stimulus. Therefore we will exclude from the analysis all the recordings contaminated with blinks or other eye movements.
Run process Events > Detect events above threshold:
Event name=blink_BAD, Channel=EEG062, All file, Maximum threshold=100, Threshold units=uV, Filter=[0.30,20.00]Hz, Use absolute value of signal.
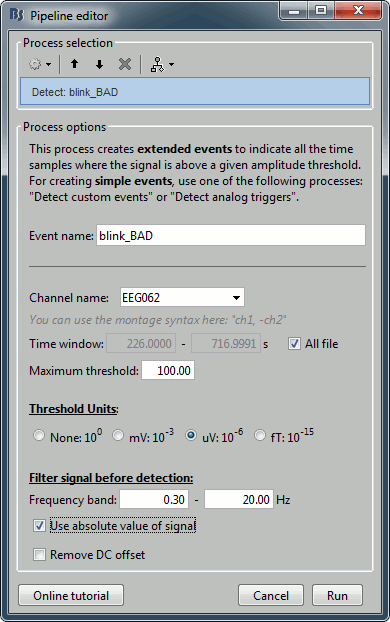
- In other tutorials, we used the process "Detect eye blinks" which creates simple events indicating the peak of the EOG signal during a blink. In this study, we preferred using this process "Detect events above threshold" because it creates extended events marking as bad all the segment for which the EOG value is above a given threshold. It's more manual but it ensures we really exclude all the ocular activity.
Inspect visually the two new cateogries of events: cardiac and blink_BAD.
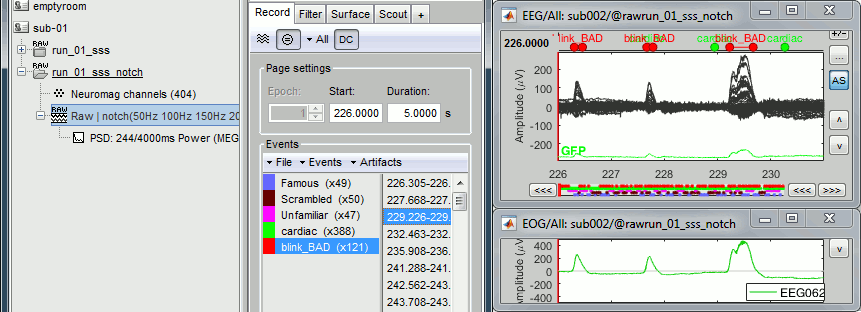
- Close all the windows (using the [X] button).
Heartbeats: Correction with SSP
- Keep the "Raw | notch" file selected in Process1.
Select process Artifacts > SSP: Heartbeats > Sensor type: MEG MAG
Add process Artifacts > SSP: Heartbeats > Sensor type: MEG GRAD - Run the execution
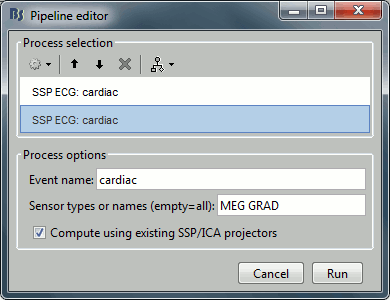
Double-click on the continuous file to show all the MEG sensors.
In the Record tab, select sensors "Left-temporal".Menu Artifacts > Select active projectors.
In category cardiac/MEG MAG: Select component #1 and view topography.
In category cardiac/MEG GRAD: Select component #1 and view topography.
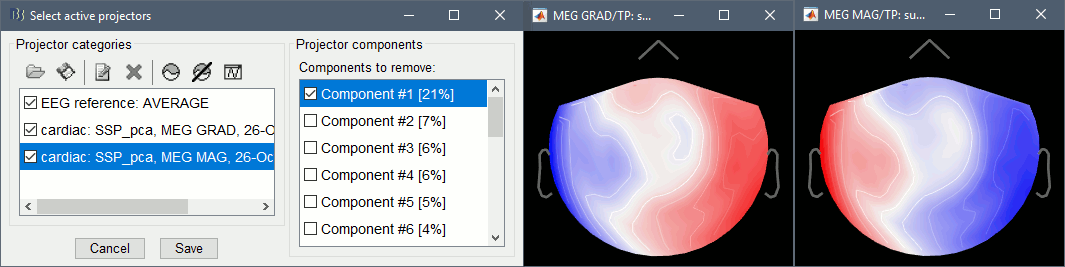
Make sure that selecting the two components removes the cardiac artifact. Then click [Save].
Additional bad segments
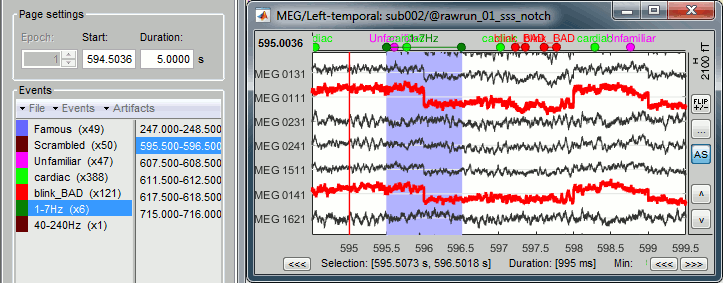
Process1>Run: Select the process "Events > Detect other artifacts". This should be done separately for MEG and EEG to avoid confusion about which sensors are involved in the artifact.
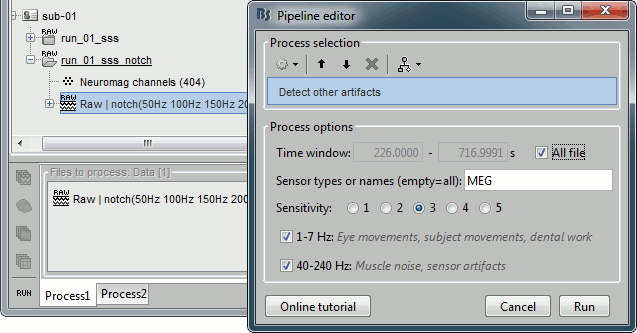
Display the MEG sensors. Review the segments that were tagged as artifact, determine if each event represents an artifact and then mark the time of the artifact as BAD. This can be done by selecting the time window around the artifact, then right-click > Reject time segment. Note that this detection process marks 1-second segments but the artifact can be shorter.
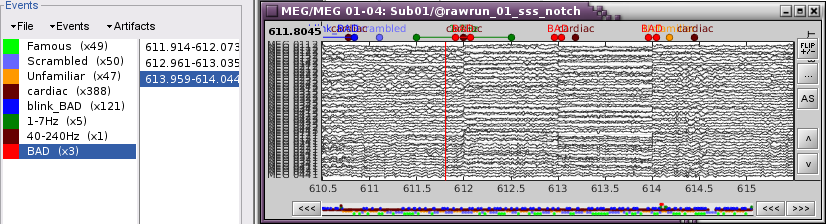
Once all the events in the two categories are reviewed and bad segments are marked, the two categories (1-7Hz and 40-240Hz) can be deleted. If want to mark as bad most of the events in a category, another solution is to add the tag BAD to its name (or menu Events > Mark group as bad), and remove from it the segments you don't want to reject.
- Do this detection and review again for the EEG.
SQUID jumps
MEG signals recorded with Elekta-Neuromag systems frequently contain SQUID jumps (more information). These sharp steps followed by a change of baseline value are easy to identify visually but more complicated to detect automatically.
The process "Detect other artifacts" usually detects most of them in the category "1-7Hz". If you observe that some are skipped, you can try re-running it with a higher sensitivity. It is important to review all the sensors and all the time in each run to be sure these events are marked as bad segments.
Epoching and averaging
Import epochs
- Keep the "Raw | notch" file selected in Process1.
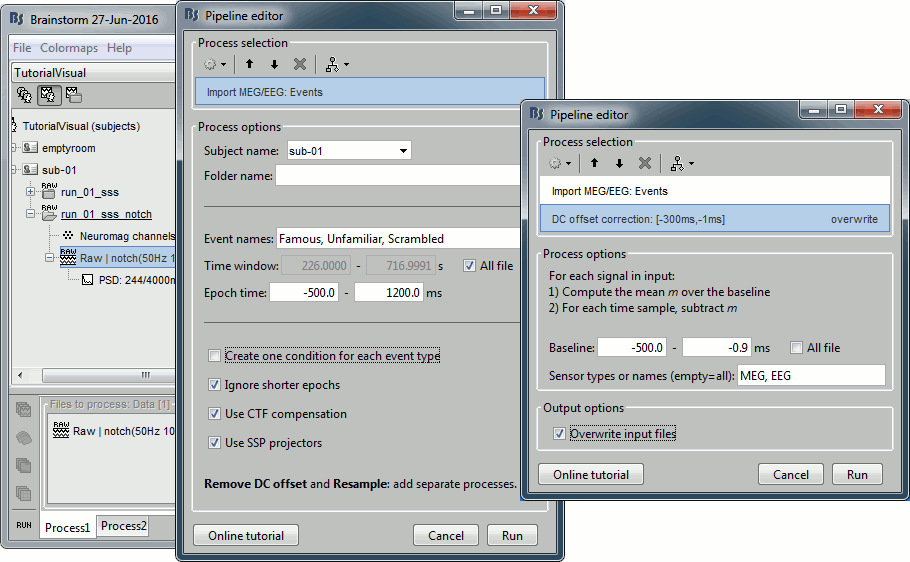
Select process: Import > Import recordings > Import MEG/EEG: Events (do not run now)
Event names "Famous, Unfamiliar, Scrambled", All file, Epoch time=[-500,1200]msAdd process: Pre-process > Remove DC offset: Baseline=[-500,-0.9]ms - Run execution.
Average by run
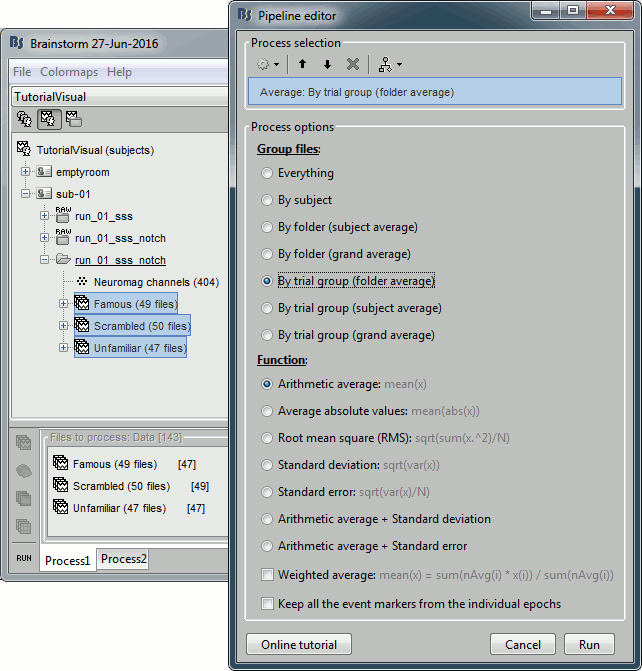
- In Process1, select all the imported trials.
Run process: Average > Average files: By trial groups (folder average)
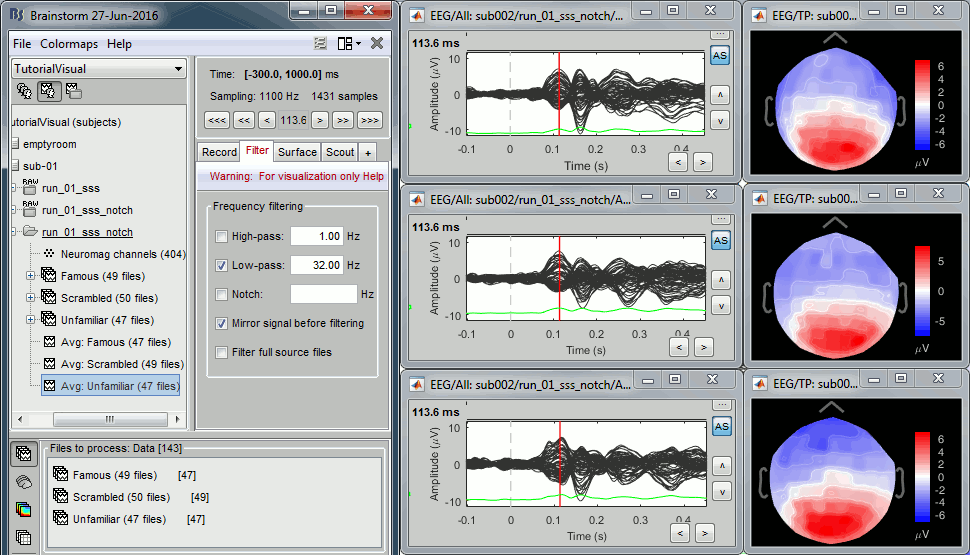
Review EEG ERP
EEG evoked response (famous, scrambled, unfamiliar), display with low-pass filter at 32Hz:
Open the Cluster tab and create a cluster with the channel EEG065 (button [NEW IND]).
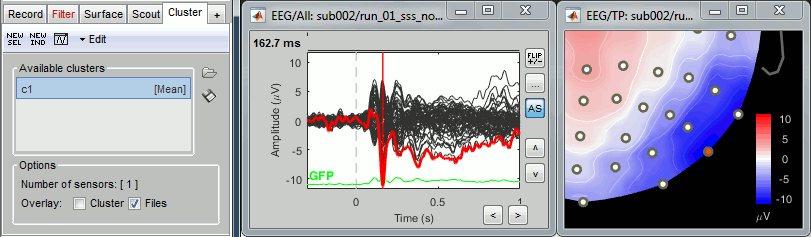
Select the cluster, select the three average files, right-click > Clusters time series (Overlay:Files).
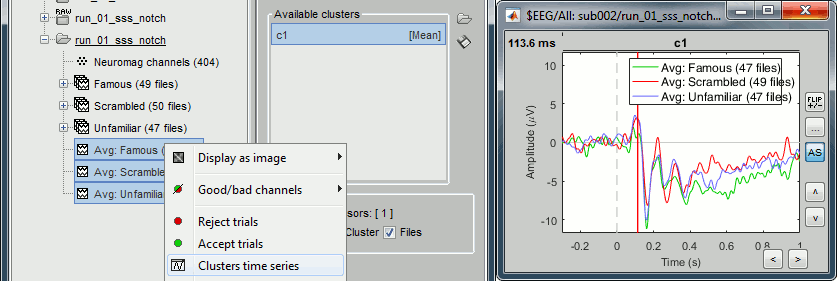
- Basic observations for EEG065 (right parieto-occipital electrode):
- Around 170ms (N170): greater negative deflection for Famous than Scrambled faces.
- After 250ms: difference between Famous and Unfamiliar faces.
Source estimation
MEG noise covariance: Empty room recordings
The minimum norm model we will use next to estimate the source activity can be improved by modeling the the noise contaminating the data. The introduction tutorials explain how to estimate the noise covariance in different ways for EEG and MEG. For the MEG recordings we will use the empty room measurements we have, and for the EEG we will compute it from the pre-stimulus baselines we have in all the imported epochs.
There are 8 empty room files available in this dataset. For each subject, we will use only one file, the one that was acquired at the closest date. We will now import and process all the empty room recordings simultaneously, even if only one is needed by the current subject. Later, for each subject we will select the most appropriate noise covariance matrix.
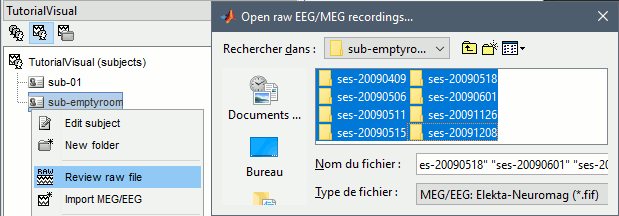
Create a new subject: sub-emptyroom
Right-click on the new subject > Review raw file.
Select all the folders in: derivatives/meg_derivatives/sub-emptyroom (processed with MaxFilter/SSS)
Do not apply default transformation, Ignore event channel.
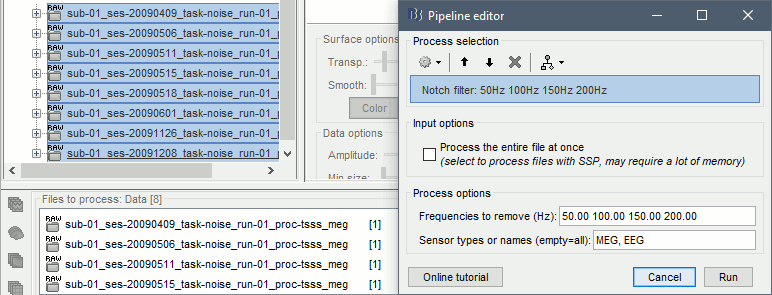
Select all these new files in Process1 and run process Pre-process > Notch filter: 50 100 150 200Hz. When using empty room measurements to compute the noise covariance, they must be processed exactly in the same way as the other recordings (same MaxFilter parameters, same frequency filters).
- Delete all the original noise files and keep only the filtered ones.
You need now to identify the noise file that was recorded the closest to the subject's recordings. In general, you can read the acquisition date of a file in the tooptip that is displayed when you leave you mouse for a while over its folder. However, the subjects' .fif files in this dataset were deidentified, we do not have access to the real acquisition dates.
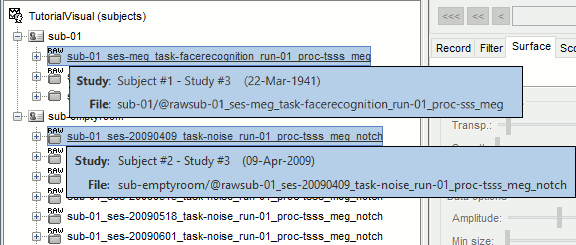
- From the BIDS meta-data, we know that the appropriate empty-room for sub-01 is ses-20090409, from April 9th 2009. We will set manually the acquisition date of all the runs in sub-01 to this same date, so that we can use the automatic matching by date in the scripts.
Right-click on the folder containing all the imported epochs > File > Set acquisition date. Enter 09-04-2009 (April 9th, 2009).
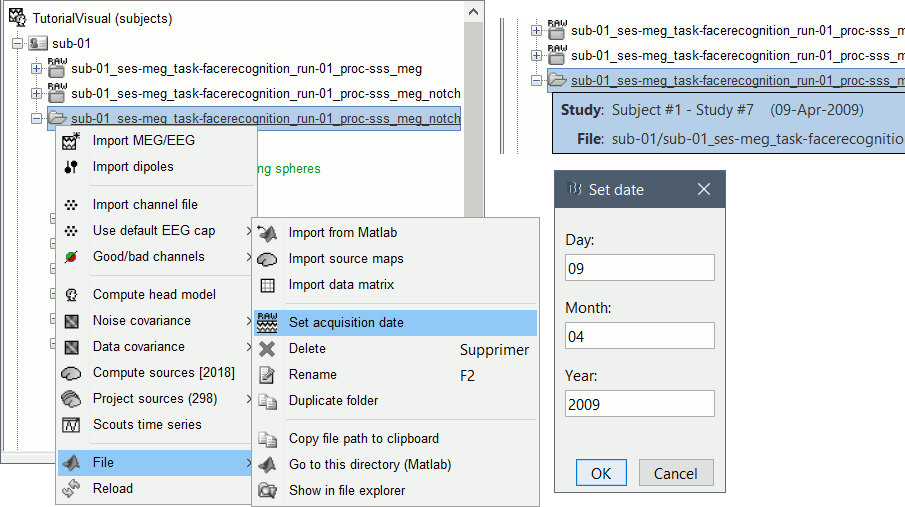
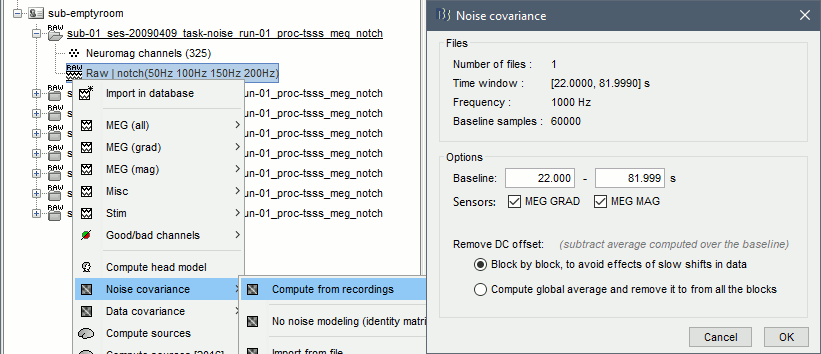
Right-click on the noise recordings 2009-04-09 > Noise covariance > Compute from recordings:
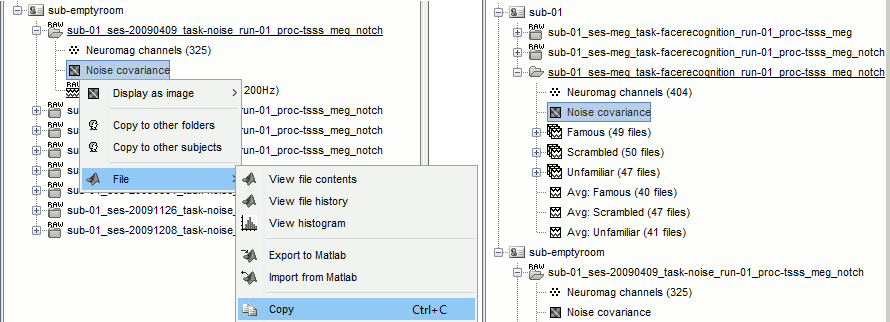
Finally copy the noise covariance from sub-emptyroom to the folder with imported epochs in sub-01. Use the popup menus File>Copy/Paste, or the keyboard shortcuts Ctrl+C/Ctrl+V.
EEG noise covariance: Pre-stimulus baseline
In folder sub-01/run-01_notch, select all the imported the imported trials, right-click > Noise covariance > Compute from recordings, Time=[-500,-0.9]ms, EEG only, Merge.
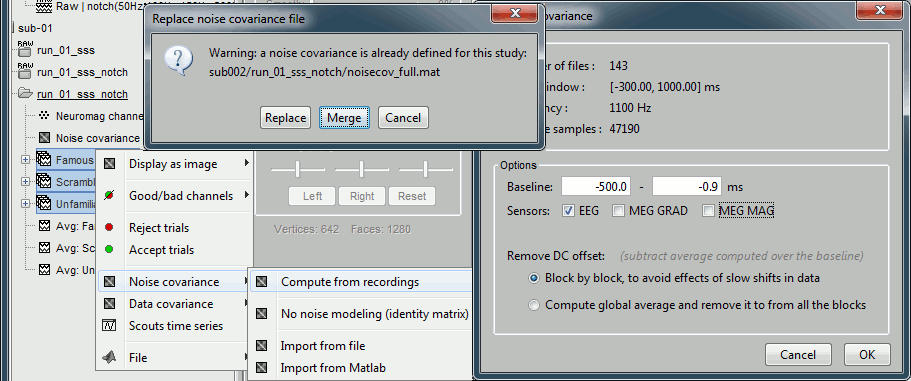
This computes the noise covariance only for EEG, and combines it with the existing MEG information.
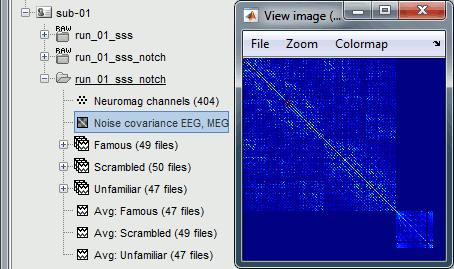
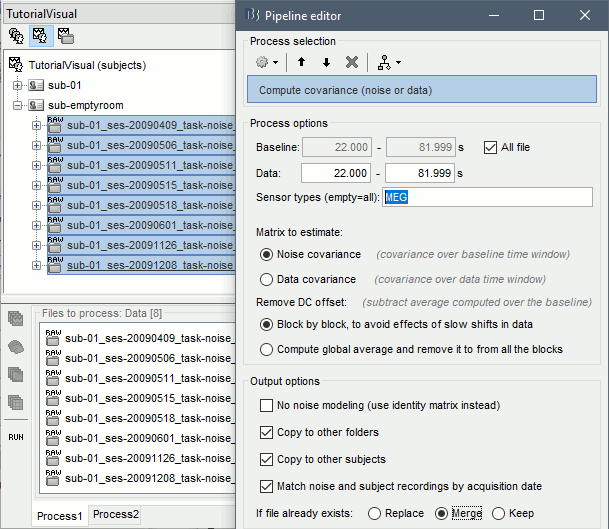
To generate the corresponding analysis script, we will use a different strategy. We will first import all the subjects, compute the EEG noise covariance for all the runs, then compute the MEG noise covariance matrix for all the empty room files at once with the process "Sources > Compute covariance" and finally merge the MEG+EEG noise covariances. When the acquisition dates are correctly set, we can use the options "Copy to other folders/subjects" and "Match noise and subject recordings by acquisition date".
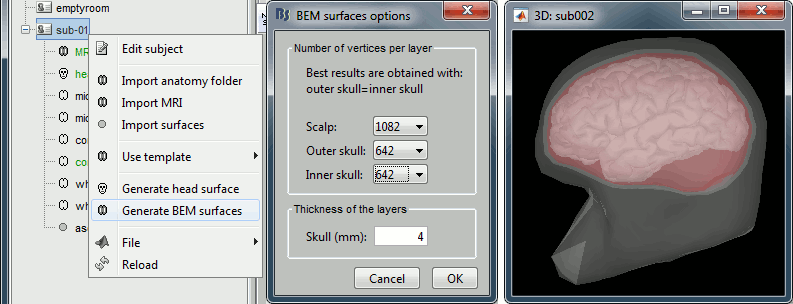
BEM layers
We will compute a BEM forward model to estimate the brain sources from the EEG recordings. For this, we need some layers defining the separation between the different tissues of the head (scalp, inner skull, outer skull).
- Go to the anatomy view (first button above the database explorer).
Right-click on the subject folder > Generate BEM surfaces: The number of vertices to use for each layer depends on your computing power and the accuracy you expect. You can try for instance with 1082 vertices (scalp) and 642 vertices (outer skull and inner skull).
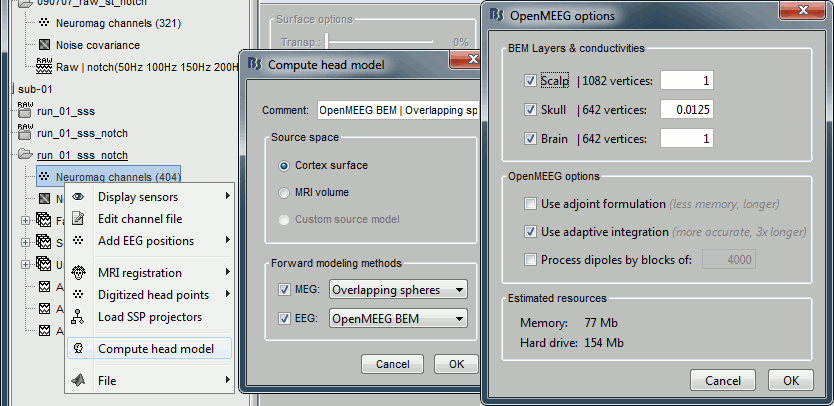
Forward model: EEG and MEG
- Go back to the functional view (second button above the database explorer).
Model used: Overlapping spheres for MEG, OpenMEEG BEM for EEG (more information).
In folder sub-01/run_01_notch, right-click on the channel file > Compute head model.
Keep all the default options . Expect this to take a while...
Inverse model: Minimum norm estimates
Right-click on head model > Compute sources [2018]: MEG MAG + GRAD (default options)
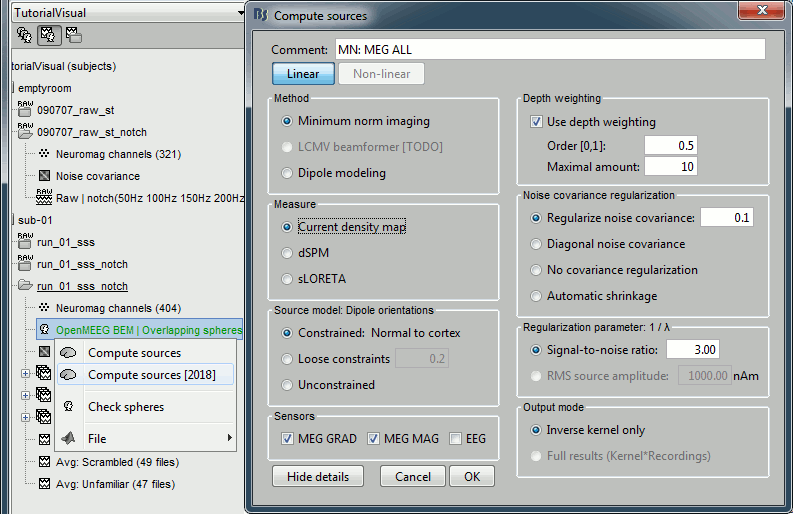
Right-click on head model > Compute sources [2018]: EEG (default bad channels).
At the end we have two inverse operators, that are shared for all the files of the run (single trials and averages). If we wanted to look at the run-level source averages, we could normalize the source maps with a Z-score wrt baseline. In this tutorial, we will first average across runs and normalize the subject-level averages. This will be done in the next tutorial (group analysis).
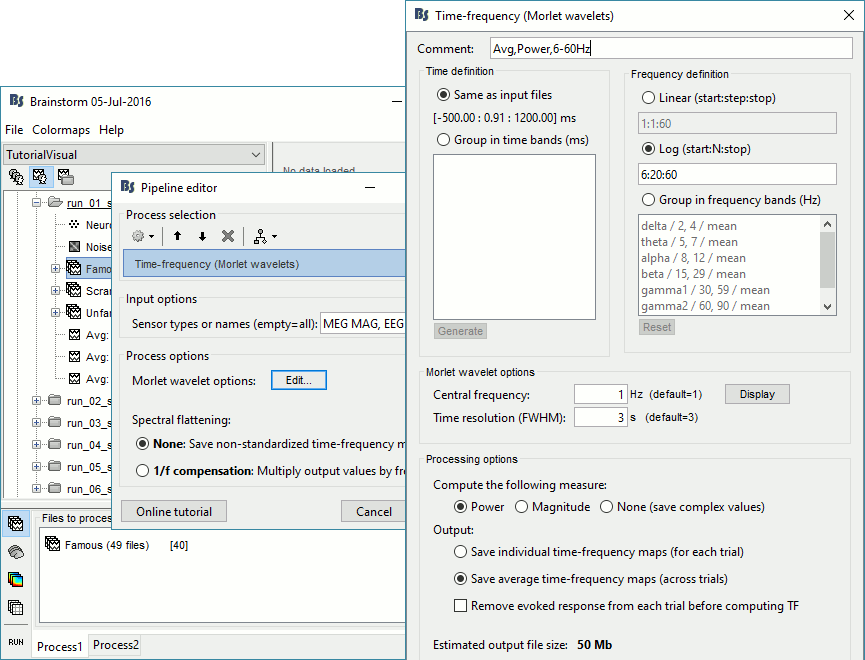
Time-frequency analysis
We will compute the time-frequency decomposition of each trial using Morlet wavelets, and average the power of the Morlet coefficients for each condition and each run separately. We will restrict the computation to the MEG magnetometers and the EEG channels to limit the computation time and disk usage.
- In Process1, select the imported trials "Famous" for run#01.
Run process Frequency > Time-frequency (Morlet wavelets): Sensor types=MEG MAG,EEG
Not normalized, Frequency=Log(6:20:60), Measure=Power, Save average
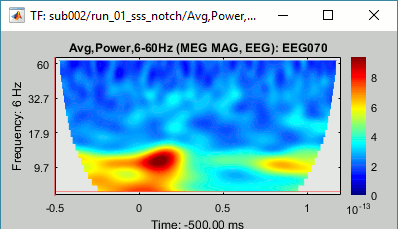
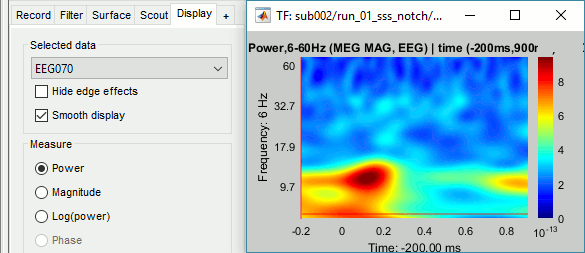
Double-click on the file to display it. In the Display tab, select the option "Hide edge effects" to exclude from the display all the values that could not be estimated in a reliable way. Let's extract only the good values from this file (-200ms to +900ms).
- In Process1, select the time-frequency file.
Run process Extract > Extract time: Time window=[-200, 900]ms, Overwrite input files
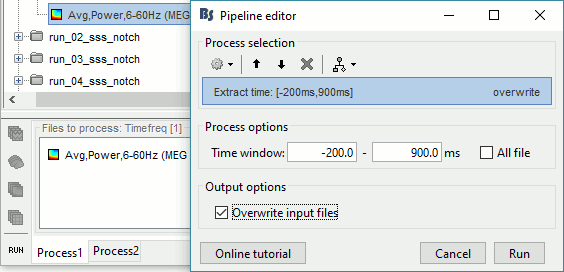
Display the file again, observe that all the possibly bad values are gone.
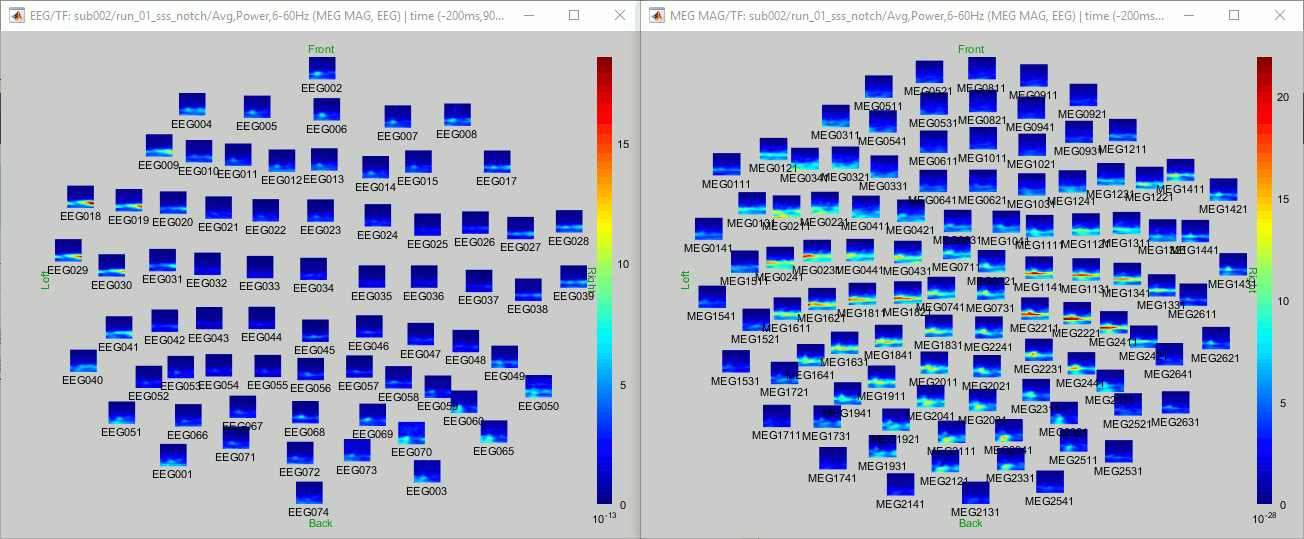
You can display all the sensors at once (MEG MAG or EEG): right-click > 2D Layout (maps).
- Repeat these steps for other conditions (Scrambled and Unfamiliar) and the other runs (2-6). There is no way with this process to compute all the averages at once, as it is with the process "Average files". This will be easier to run from a script.
- If we wanted to look at the run-level source averages, we could normalize these time-frequency maps. In this tutorial, we will first average across runs and normalize the subject-level averages. This will be done in the next tutorial (group analysis).
Scripting
We have now all the files we need for the group analysis (next tutorial). We need to repeat the same operations for all the runs and all the subjects. Some of these steps are fully automatic and take a lot of time (filtering, computing the forward model), they should be executed from a script.
However, we recommend you always review manually some of the pre-processing steps (selection of the bad segments and bad channels, SSP/ICA components). Do not trust blindly any fully automated cleaning procedure.
For the strict reproducibility of this analysis, we provide a script that processes all the 16 subjects: brainstorm3/toolbox/script/tutorial_visual_single.m (execution time: 10-30 hours)
Report for the first subject: report_TutorialVisual_sub-01.html
You should note that this is not the result of a fully automated procedure. The bad channels were identified manually and are defined for each run in the script. The bad segments were detected automatically, confirmed manually for each run and saved in external files, then exported as text ans copied at the end of this script.
All the process calls (bst_process) were generated automatically using with the script generator (menu Generate .m script in the pipeline editor). Everything else was added manually (loops, bad channels, file copies).