MEG visual tutorial: Single subject
Authors: Francois Tadel, Elizabeth Bock.
The aim of this tutorial is to reproduce in the Brainstorm environment the analysis described in the SPM tutorial "Multimodal, Multisubject data fusion". The data processed here consists in simultaneous MEG/EEG recordings of 19 subjects performing simple visual task on a large number of famous, unfamiliar and scrambled faces.
The analysis is split in two tutorial pages: the present tutorial describes the detailed analysis of one single subject and another one that describes the batch processing and group analysis of the 19 subjects.
Note that the operations used here are not detailed, the goal of this tutorial is not to teach Brainstorm to a new inexperienced user. For in depth explanations of the interface and the theory, please refer to the introduction tutorials.
Contents
License
This dataset was obtained from the OpenfMRI project (http://www.openfmri.org), accession #ds117. It is made available under the Creative Commons Attribution 4.0 International Public License. Please cite the following reference if you use these data:
Wakeman DG, Henson RN, A multi-subject, multi-modal human neuroimaging dataset, Scientific Data (2015)
Any questions, please contact: rik.henson@mrc-cbu.cam.ac.uk
Presentation of the experiment
Experiment
19 subjects (three are excluded from the group analysis for various reasons)
- 6 acquisition runs (aka sessions) of approximately 10mins for each subject
- Presentation of series of images: familiar faces, unfamiliar faces, phase-scrambled faces
- The subject has to judge the left-right symmetry of each stimulus
- Total of nearly 300 trials for each of the 3 conditions
MEG acquisition
Acquisition at 1100Hz with an Elekta-Neuromag VectorView system (simultaneous MEG+EEG).
- Recorded channels (404):
- 102 magnetometers
- 204 planar gradiometers
- 70 EEG electrodes recorded with a nose reference.
MEG data have been "cleaned" using Signal-Space Separation as implemented in MaxFilter 2.2.
- A Polhemus device was used to digitize three fiducial points and a large number of other points across the scalp, which can be used to coregister the M/EEG data with the structural MRI image.
- Stimulation triggers: The triggers related with the visual presentation are saved in the STI101 channel, with the following event codes (bit 3 = face, bit 4 = unfamiliar, bit 5 = scrambled):
- Famous faces: 5 (00101), 6 (00110), 7 (00111)
- Unfamiliar faces: 13 (01101), 14 (01110), 15 (01111)
- Scrambled images: 17 (10001), 18 (10010), 19 (10011)
Delays between the trigger in STI101 and the actual presentation of stimulus: 34.5ms
The distribution includes MEG noise recordings acquired around the dates of the experiment, processed with MaxFilter 2.2 in the same way as the experimental data.
Subject anatomy
- MRI data acquired on a 3T Siemens TIM Trio: 1x1x1mm T1-weighted structural MRI.
- The face has been removed from the strucural images for anonymization purposes.
Processed with FreeSurfer 5.3.
Download and installation
First, make sure you have enough space on your hard drive, at least 350Gb:
The downloaded raw files: 100Gb
The database after processing: 250Gb
The data is hosted on the OpenfMRI website: https://openfmri.org/dataset/ds000117/
Download all the files available from this website (approximately 160Gb):
ds117_metadata.tgz, ds117_sub001_raw.tgz, ..., ds117_sub019_raw.tgz- Unzip all the .tgz files in the same folder.
The FreeSurfer segmentations of the T1 images are not part of the OpenfMRI distribution. You can either process them by yourself, or download the result of the segmentation from the Brainstorm website.
Go to the Download page, and download the file: sample_group_freesurfer.zip
Unzip this file in the same folder as the other files.- We will use the following files from this distribution:
/anatomy/subXXX/: Segmentation folders generated with FreeSurfer.
/emptyroom/090707_raw_st.fif: MEG empty room measurements, processed with MaxFilter.
/ds117/subXXX/MEG/*_sss.fif: MEG and EEG recordings, processed with MaxFilter.
/README: License and dataset description.
- Reminder: Do not put the downloaded files in the Brainstorm folders (program or database folders).
Start Brainstorm (Matlab scripts or stand-alone version). For help, see the Installation page.
Select the menu File > Create new protocol. Name it "TutorialVisual" and select the options:
"No, use individual anatomy",
"No, use one channel file per condition".
Import the anatomy
This page explains how to import and process subject #002 only. Subject #001 was not the best example because it will be later excluded from the EEG group analysis (incorrect electrodes positions).
- Switch to the "anatomy" view.
Right-click on the TutorialVisual folder > New subject > sub002
- Leave the default options you defined for the protocol.
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: anatomy/freesurfer/sub002 (from sample_group_freesurfer.zip)
- Number of vertices of the cortex surface: 15000 (default value)
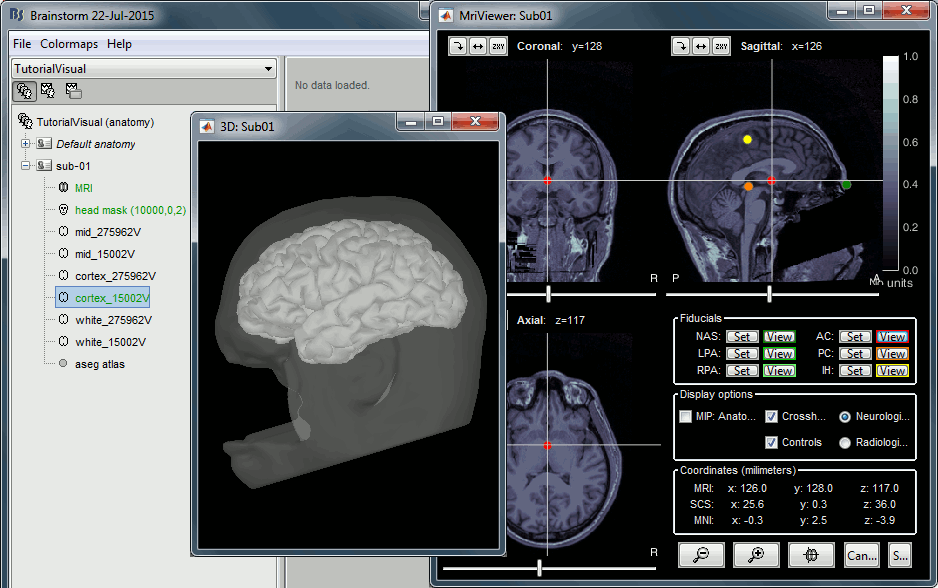
- The two sets of fiducials we usually have to define interactively are here automatically set.
NAS/LPA/RPA: The file Anatomy/Sub01/fiducials.m contains the definition of the nasion, left and right ears. The anatomical points used by the authors are the same as the ones we recommend in the Brainstorm coordinates systems page.
AC/PC/IH: Automatically identified using the SPM affine registration with an MNI template.
If you want to double-check that all these points were correctly marked after importing the anatomy, right-click on the MRI > Edit MRI.
At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface. Do not worry about the big holes in the head surface, parts of MRI have been remove voluntarily for anonymization purposes.
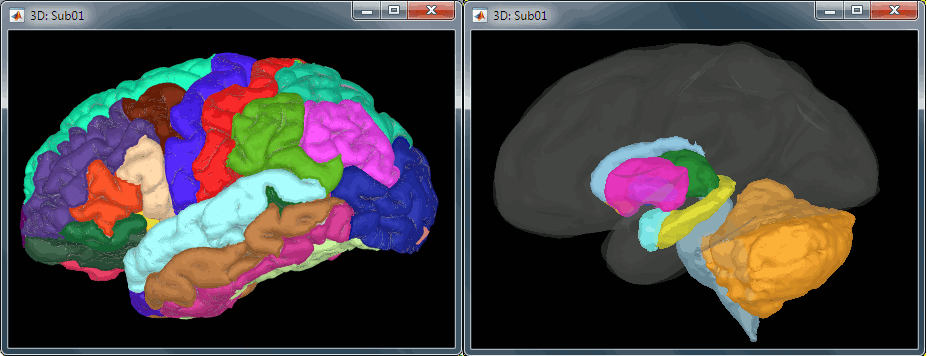
All the anatomical atlases generated by FreeSurfer were automatically imported: the cortical atlases (Desikan-Killiany, Mindboggle, Destrieux, Brodmann) and the sub-cortical regions (ASEG atlas).
Access the recordings
Link the recordings
We need to attach the continuous .fif files containing the recordings to the database.
- Switch to the "functional data" view.
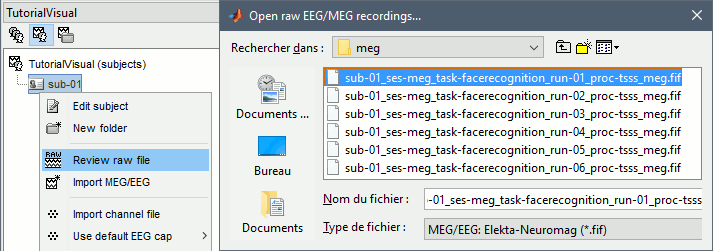
Right-click on the subject folder > Review raw file.
Select the file format: "MEG/EEG: Neuromag FIFF (*.fif)"
Select the first file in the MEG folder for subject 002: sub002/MEEG/run_01_sss.fif
Events: Ignore. We will read the stimulus triggers later.
Refine registration now? NO
The head points that are available in the FIF files contain all the points that were digitized during the MEG acquisition, including the ones corresponding to the parts of the face that have been removed from the MRI. If we run the fitting algorithm, all the points around the nose will not match any close points on the head surface, leading to a wrong result. We will first remove the face points and then run the registration manually.
Channel classification
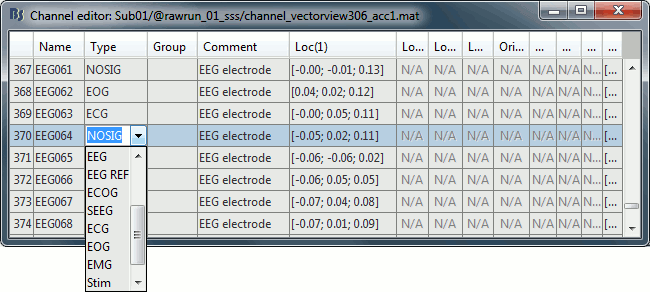
A few non-EEG channels are mixed in with the EEG channels, we need to change this before applying any operation on the EEG channels.
Right-click on the channel file > Edit channel file. Double-click on a cell to edit it.
Change the type of EEG062 to EOG (electrooculogram).
Change the type of EEG063 to ECG (electrocardiogram).
Change the type of EEG061 and EEG064 to NOSIG. Close the window and save the modifications.
MRI registration
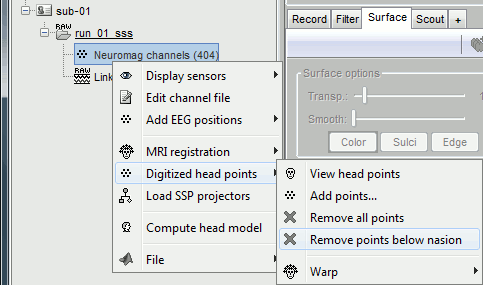
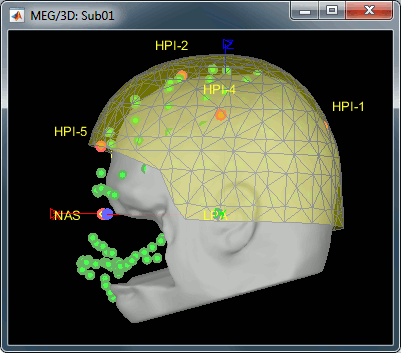
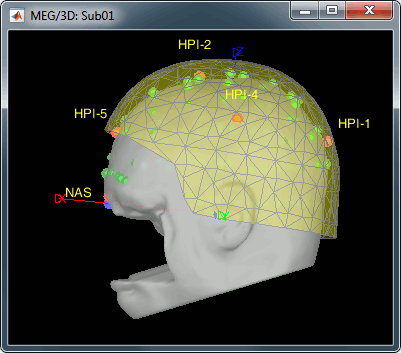
At this point, the registration MEG/MRI is based only on the three anatomical landmarks NAS/LPA/RPA. All the MRI scans were anonymized (defaced) and for some subjects the nasion could not be defined properly. We will try to refine this registration using the additional head points that were digitized (only the points above the nasion).
Right-click on the channel file > Digitized head points > Remove points below nasion.
Right-click on the channel file > MRI registration > Refine using head points.
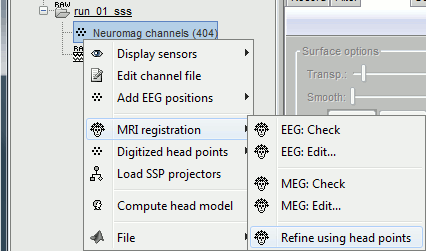
MEG/MRI registration, before (left) and after (right) this automatic registration procedure:
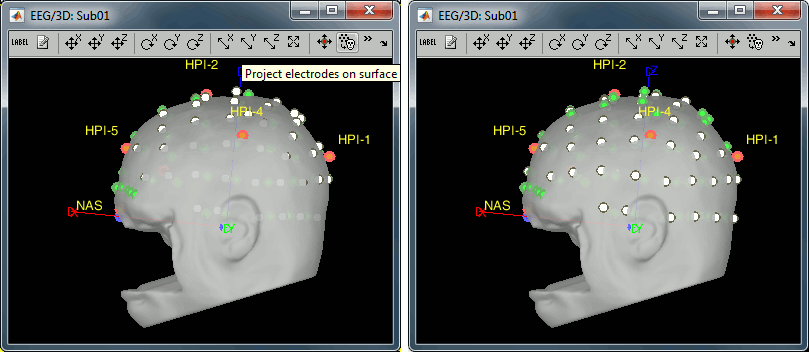
Right-click on the channel file > MRI registration > EEG: Edit...
Click on [Project electrodes on surface], then close the figure to save the modifications.
Read stimulus triggers
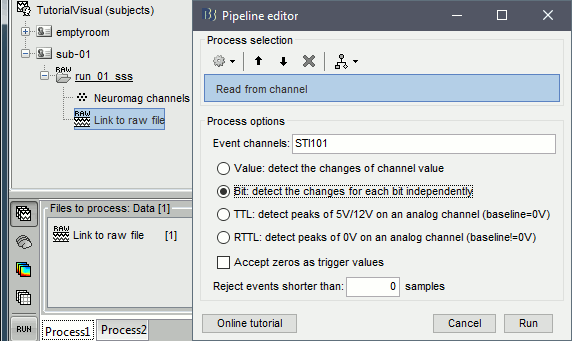
We need to read the stimulus markers from the STI channels. The following tasks can be done in an interactive way with menus in the Record tab, as in the introduction tutorials. We will illustrate here how to do this with the pipeline editor, it will be easier to batch it for all the runs and all the subjects.
- In Process1, select the "Link to raw file", click on [Run].
Select process Events > Read from channel, Channel: STI101, Detection mode: Bit.
Do not execute the process, we will add other processes to classify the markers.
- We want to create three categories of events, based on their numerical codes:
Famous faces: 5 (00101), 6 (00110), 7 (00111) => Bit 3 only
Unfamiliar faces: 13 (01101), 14 (01110), 15 (01111) => Bit 3 and 4
Scrambled images: 17 (10001), 18 (10010), 19 (10011) => Bit 5 only
- We will start by creating the category "Unfamiliar" (combination of events "3" and "4") and remove the remove the initial events. Then we just have to rename the remaining "3" in "Famous", and all the "5" in "Scrambled".
Add process Events > Group by name: "Unfamiliar=3,4", Delay=0, Delete original events
Add process Events > Rename event: 3 => Famous
Add process Events > Rename event: 5 => Scrambled
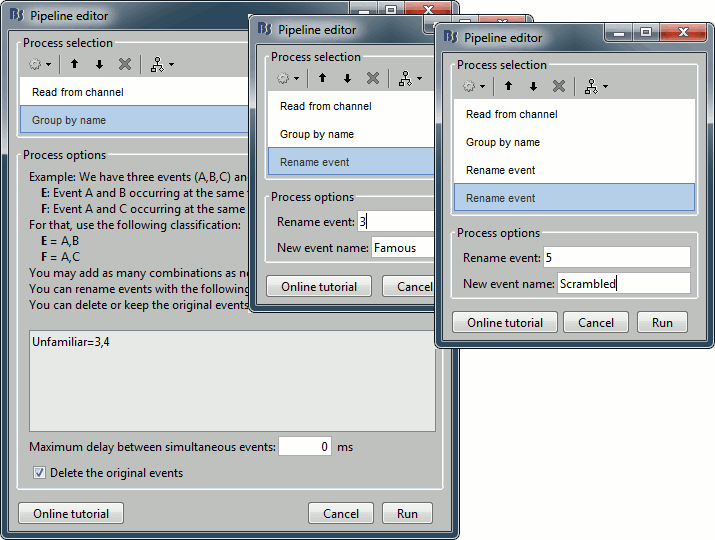
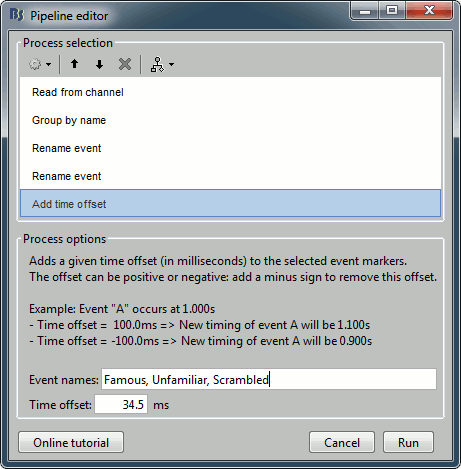
Add process Events > Add time offset to correct for the presentation delays:
Event names: "Famous, Unfamiliar, Scrambled", Time offset = 34.5ms
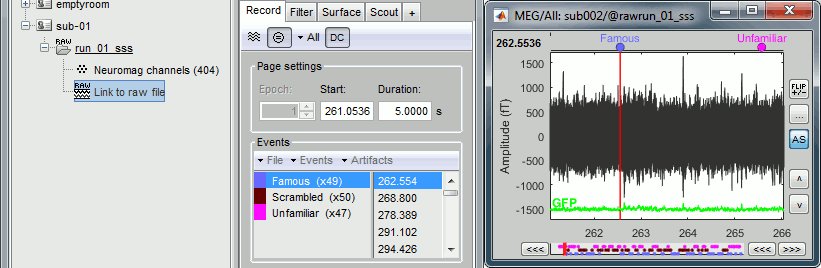
Finally run the script. Double-click on the recordings to make sure the labels were detected correctly. You can delete the unwanted events that are left in the recordings (1,2,9,13):
Pre-processing
Spectral evaluation
- Keep the "Link to raw file" in Process1.
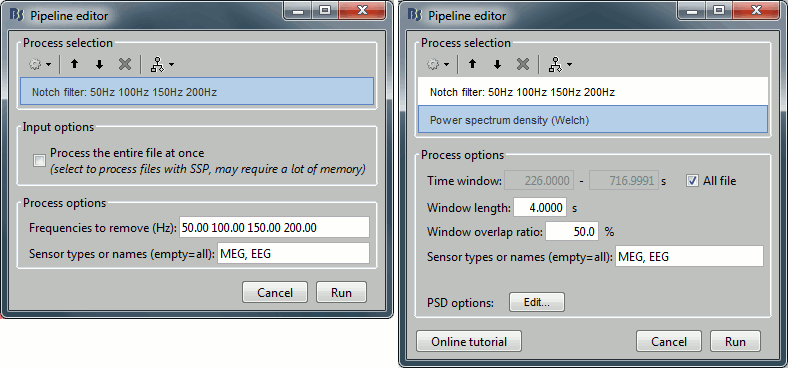
Run process Frequency > Power spectrum density (Welch) with the options illustrated below.
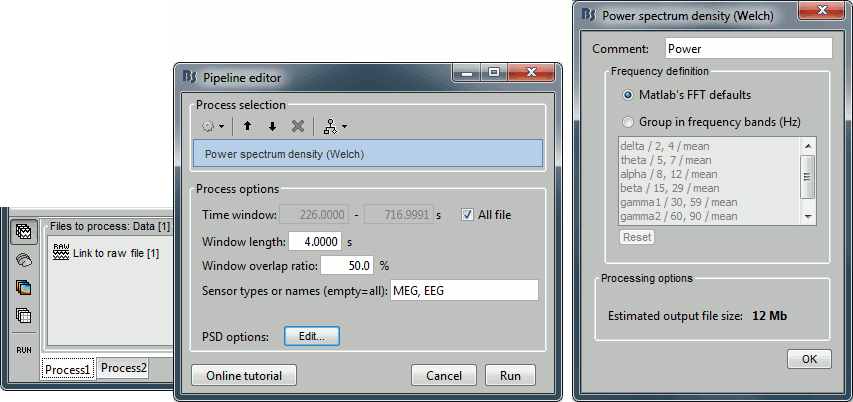
Right-click on the PSD file > Power spectrum.
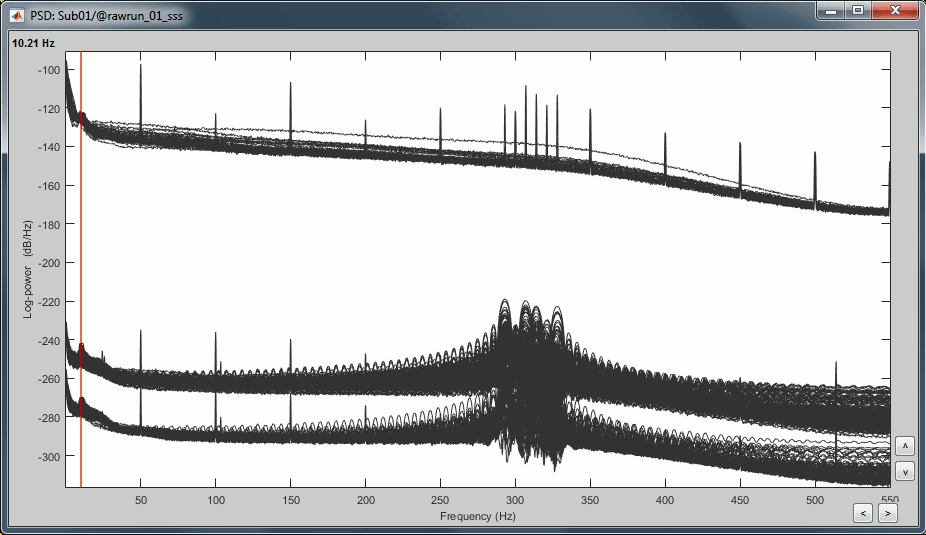
- Observations:
- Three groups of sensors, from top to bottom: EEG, MEG gradiometers, MEG magnetometers.
Power lines: 50 Hz and harmonics
- Alpha peak around 10 Hz
Artifacts due to Elekta electronics: 293Hz, 307Hz, 314Hz, 321Hz, 328Hz.
Suspected bad EEG channels: EEG016
- Close all the windows.
Remove line noise
- Keep the "Link to raw file" in Process1.
Select process Pre-process > Notch filter to remove the line noise (50-200Hz).
Add the process Frequency > Power spectrum density (Welch).
Double-click on the PSD for the new continuous file to evaluate the quality of the correction.
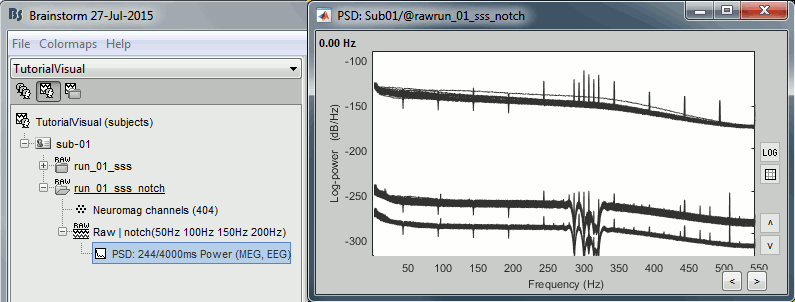
- Close all the windows (use the [X] button at the top-right corner of the Brainstorm window).
EEG reference and bad channels
Right-click on link to the processed file ("Raw | notch(50Hz ...") > EEG > Display time series.
Select channel EEG016 and mark it as bad (using the popup menu or pressing the Delete key).
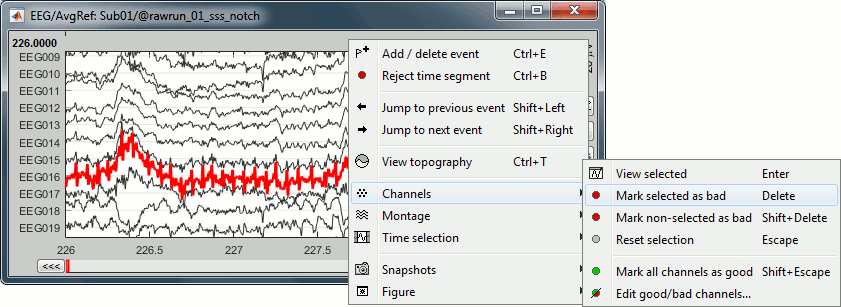
In the Record tab, menu Artifacts > Re-reference EEG > "AVERAGE".
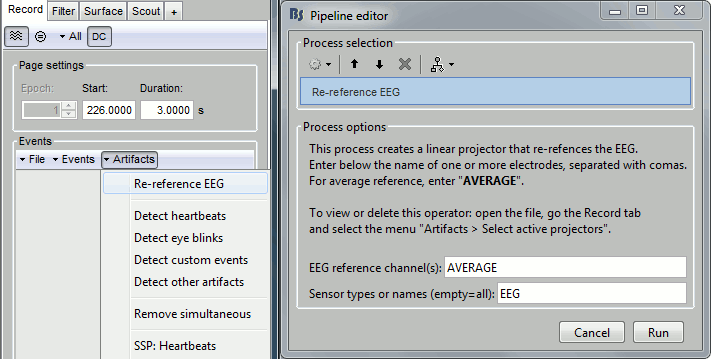
At the end, the window "select active projectors" is open to show the new re-referencing projector. Just close this window. To get it back, use the menu Artifacts > Select active projectors.
Artifact detection
Heartbeats: Detection
Empty the Process1 list (right-click > Clear list).
- Drag and drop the continuous processed file ("Raw | notch(50Hz...)") to the Process1 list.
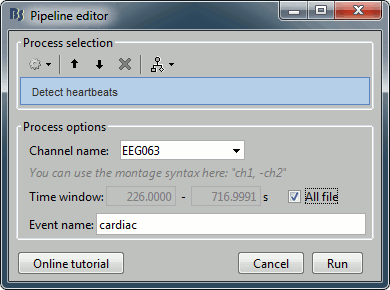
Run process Events > Detect heartbeats: Channel name=EEG063, All file, Event name=cardiac
- In many of the other tutorials, we detect the blinks and remove them with SSP. In this experiment, we are particularly interested in the subject's response to seeing the stimulus. Therefore we will exclude from the analysis all the recordings contaminated with blinks or other eye movements.
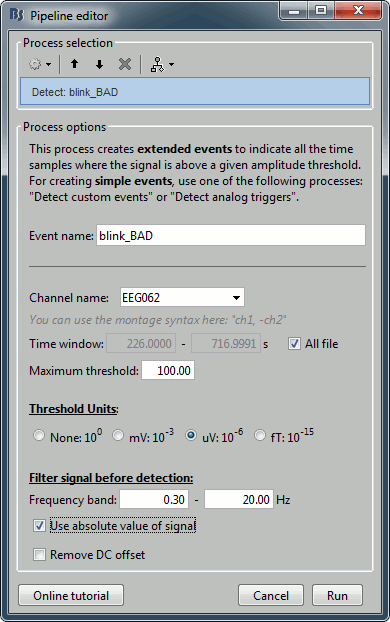
Run process Artifacts > Detect events above threshold:
Event name=blink_BAD, Channel=EEG062, All file, Maximum threshold=100, Threshold units=uV, Filter=[0.30,20.00]Hz, Use absolute value of signal.
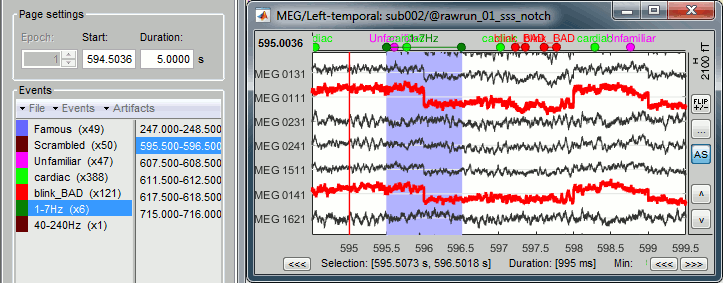
Inspect visually the two new cateogries of events: cardiac and blink_BAD.
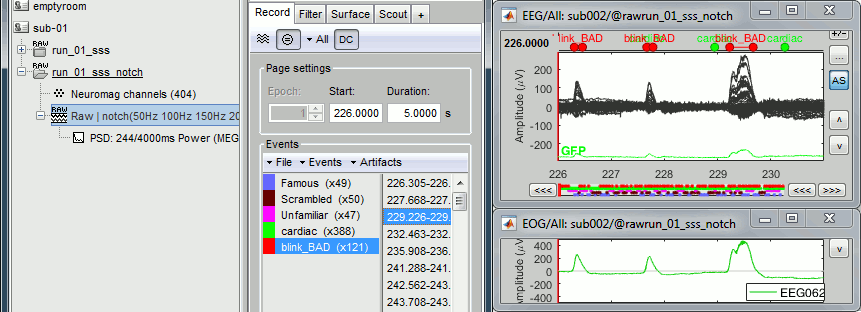
- Close all the windows (using the [X] button).
Heartbeats: Correction with SSP
- Keep the "Raw | notch" file selected in Process1.
Select process Artifacts > SSP: Heartbeats > Sensor type: MEG MAG
Add process Artifacts > SSP: Heartbeats > Sensor type: MEG GRAD - Run the execution
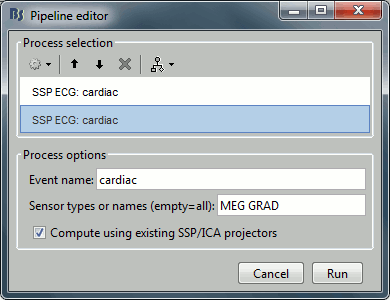
Double-click on the continuous file to show all the MEG sensors.
In the Record tab, select sensors "Left-temporal".Menu Artifacts > Select active projectors.
In category cardiac/MEG MAG: Select component #1 and view topography.
In category cardiac/MEG GRAD: Select component #1 and view topography.
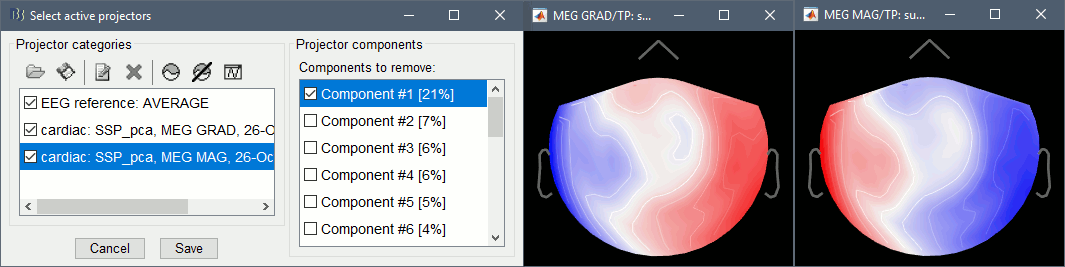
Make sure that selecting the two components removes the cardiac artifact. Then click [Save].
Additional bad segments
Process1>Run: Select the process "Events > Detect other artifacts". This should be done separately for MEG and EEG to avoid confusion about which sensors are involved in the artifact.
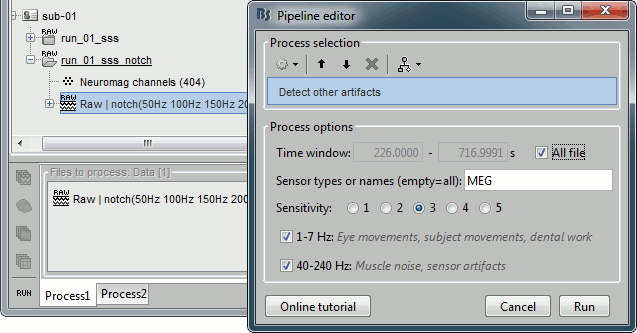
Display the MEG sensors. Review the segments that were tagged as artifact, determine if each event represents an artifact and then mark the time of the artifact as BAD. This can be done by selecting the time window around the artifact, then right-click > Reject time segment. Note that this detection process marks 1-second segments but the artifact can be shorter.
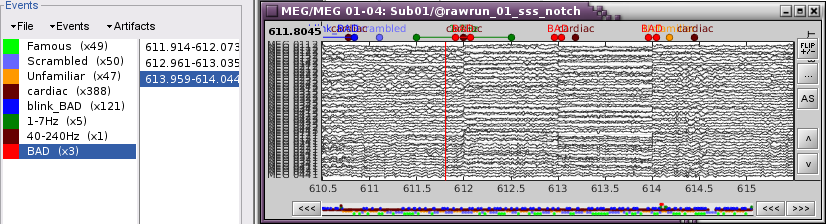
- Once all the events in the two categories are reviewed and bad segments are marked, the two categories (1-7Hz and 40-240Hz) can be deleted.
- Do this detection and review again for the EEG.
SQUID jumps
MEG signals recorded with Elekta-Neuromag systems frequently contain SQUID jumps (more information). These sharp steps followed by a change of baseline value are easy to identify visually but more complicated to detect automatically.
The process "Detect other artifacts" usually detects most of them in the category "1-7Hz". If you observe that some are skipped, you can try re-running it with a higher sensitivity. It is important to review all the sensors and all the time in each run to be sure these events are marked as bad segments.
Epoching and averaging
Import epochs
- Keep the "Raw | notch" file selected in Process1.
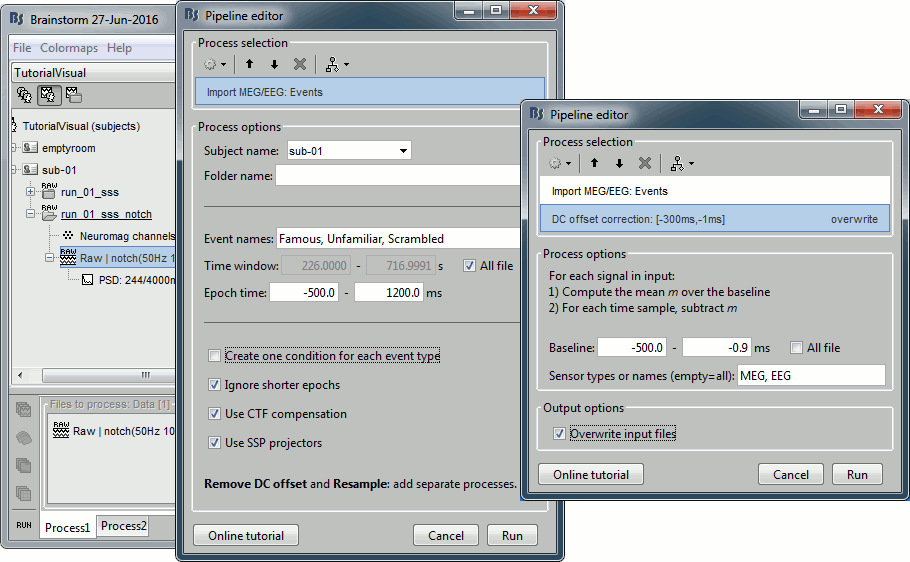
Select process: Import recordings > Import MEG/EEG: Events (do not run immediately)
Event names "Famous, Unfamiliar, Scrambled", All file, Epoch time=[-500,1200]msAdd process: Pre-process > Remove DC offset: Baseline=[-500,-0.9]ms - Run execution.
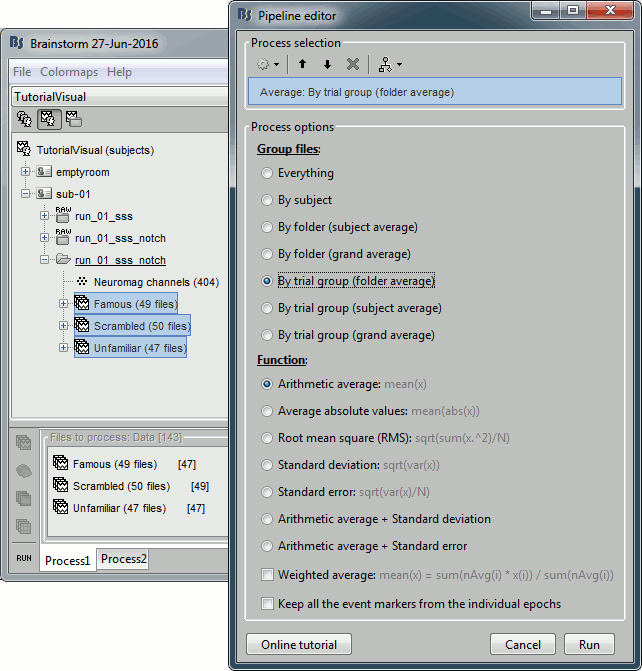
Average by run
- In Process1, select all the imported trials.
Run process: Average > Average files: By trial groups (folder average)
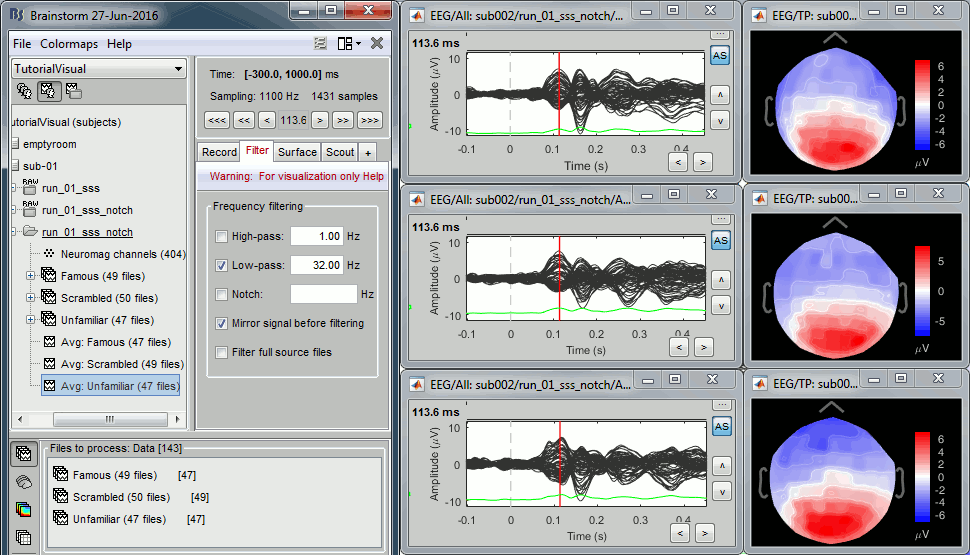
Review EEG ERP
EEG evoked response (famous, scrambled, unfamiliar):
Open the Cluster tab and create a cluster with the channel EEG065 (button [NEW IND]).
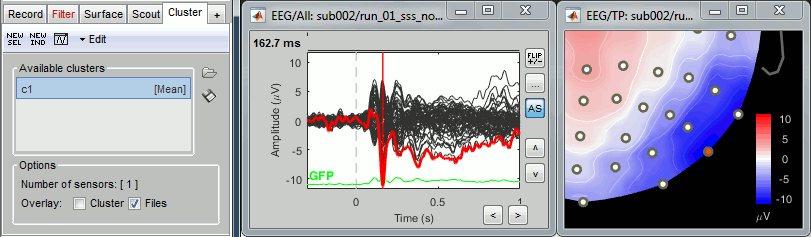
Select the cluster, select the three average files, right-click > Clusters time series (Overlay:Files).
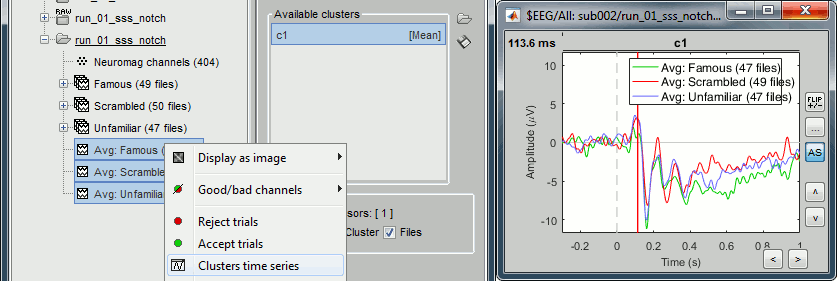
- Basic observations for EEG065 (right parieto-occipital electrode):
- Around 170ms (N170): greater negative deflection for Famous than Scrambled faces.
- After 250ms: difference between Famous and Unfamiliar faces.
Source estimation
MEG noise covariance: Empty room recordings
The minimum norm model we will use next to estimate the source activity can be improved by modeling the the noise contaminating the data. The section shows how to estimate the noise covariance in different ways for EEG and MEG. For the MEG recordings we will use the empty room measurements we have, and for the EEG we will compute it from the pre-stimulus baselines we have in all the imported epochs.
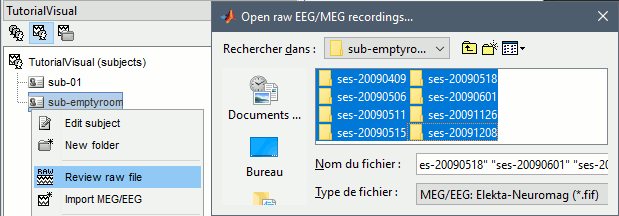
Create a new subject: emptyroom
Right-click on the new subject > Review raw file.
Select file: sample_group/emptyroom/090707_raw_st.fif
Do not apply default transformation, Ignore event channel.
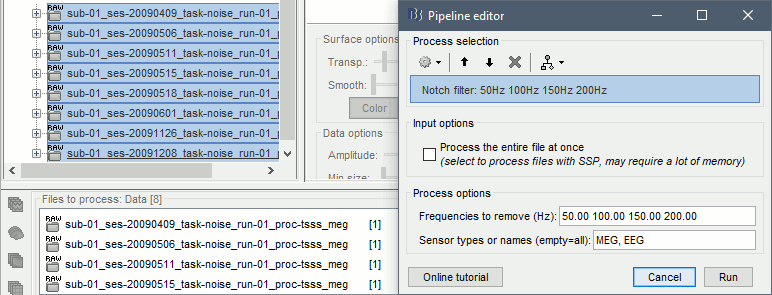
Select this new file in Process1 and run process Pre-process > Notch filter: 50 100 150 200Hz. When using empty room measurements to compute the noise covariance, they must be processed exactly in the same way as the other recordings.
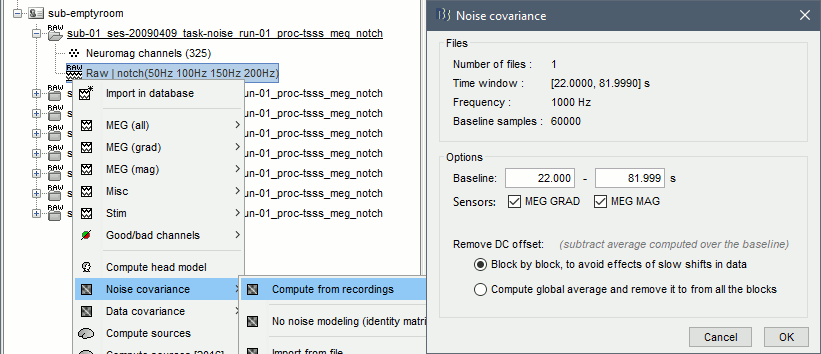
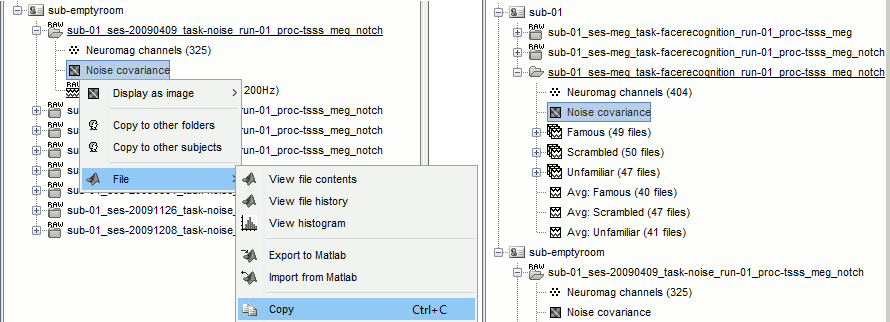
Right-click on the filtered noise recordings > Noise covariance > Compute from recordings:
Right-click on the Noise covariance > Copy to other subjects
EEG noise covariance: Pre-stimulus baseline
In folder sub002/run_01_sss_notch, select all the imported the imported trials, right-click > Noise covariance > Compute from recordings, Time=[-500,-0.9]ms, EEG only, Merge.
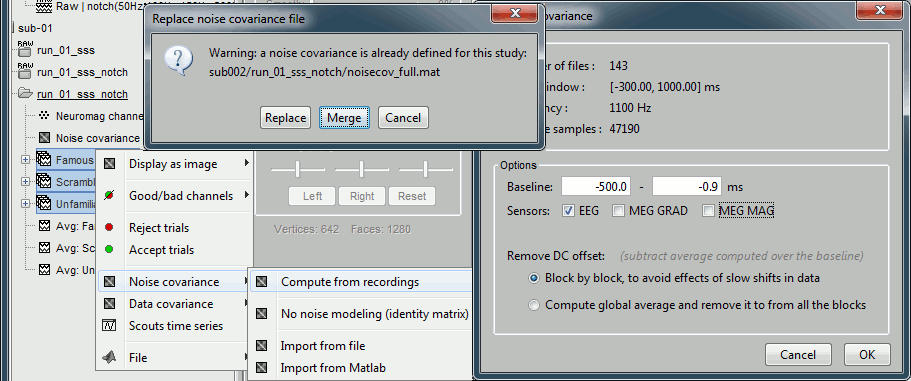
This computes the noise covariance only for EEG, and combines it with the existing MEG information.
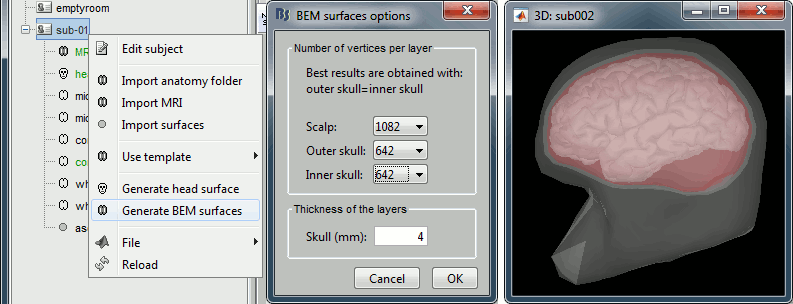
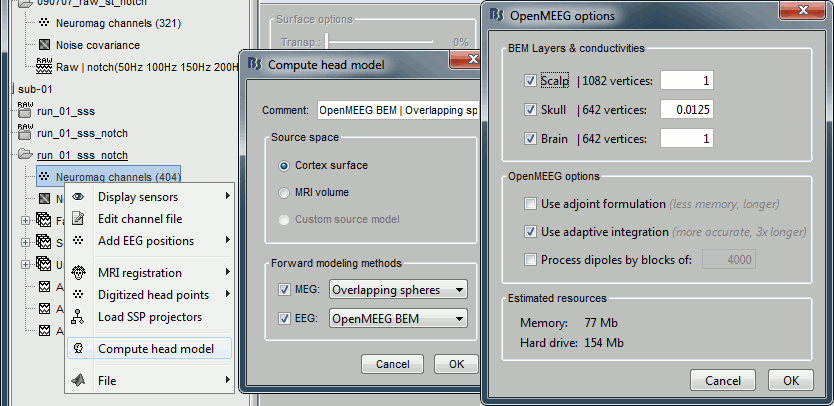
BEM layers
We will compute a BEM forward model to estimate the brain sources from the EEG recordings. For this, we need some layers defining the separation between the different tissues of the head (scalp, inner skull, outer skull).
- Go to the anatomy view (first button above the database explorer).
Right-click on the subject folder > Generate BEM surfaces: The number of vertices to use for each layer depends on your computing power and the accuracy you expect. You can try for instance with 1082 vertices (scalp) and 642 vertices (outer skull and inner skull).
Forward model: EEG and MEG
- Go back to the functional view (second button above the database explorer).
Model used: Overlapping spheres for MEG, OpenMEEG BEM for EEG (more information).
In folder sub002/run_01_sss_notch, right-click on the channel file > Compute head model.
Keep all the default options . Expect this to take a while...
Inverse model: Minimum norm estimates
Right-click on the new head model > Compute sources [2016]: MEG MAG + GRAD (default options)
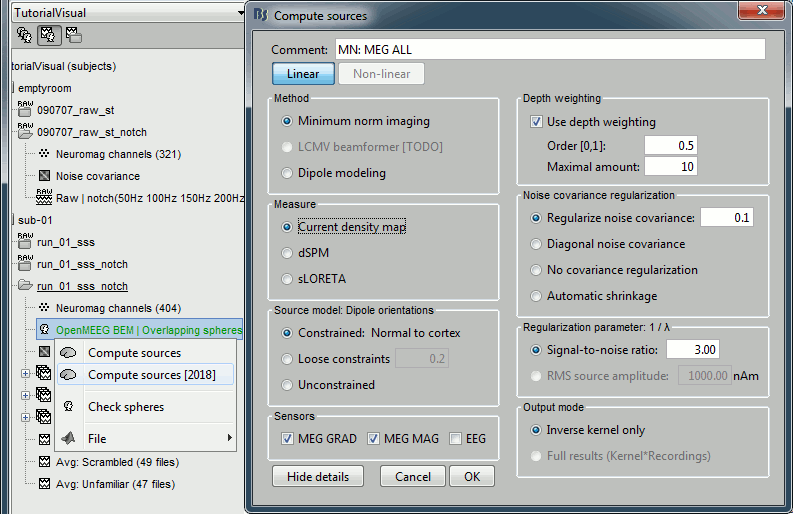
Right-click on the new head model > Compute sources [2016]: EEG (default bad channels)
Scripting
We have now all the files we need for the group analysis (next tutorial). We need to repeat the same operations for all the runs and all the subjects. Some of these steps are fully automatic and take a lot of time (filtering, computing the forward model), they should be executed from a script.
However, we recommend you always review manually some of the pre-processing steps (selection of the bad segments and bad channels, SSP/ICA components). Do not trust blindly any fully automated cleaning procedure.
For the strict reproducibility of this analysis, we provide a script that processes all the 19 subjects: brainstorm3/toolbox/script/tutorial_visual_single.m
You should note that this is not the result of a fully automated procedure. The bad channels were identified manually and are defined for each run in the script. The bad segments were detected automatically, confirmed manually for each run and saved in external files distributed with the Brainstorm package sample_group_freesurfer.zip (sample_group/brainstorm/bad_segments/*.mat).
All the process calls (bst_process) were generated automatically using with the script generator (menu Generate .m script in the pipeline editor). Everything else was added manually (loops, bad channels, file copies).