|
Size: 32989
Comment:
|
Size: 42966
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| = MEG corticomuscular coherence = | = Corticomuscular coherence (MEG) = |
| Line 6: | Line 6: |
| ''Authors: Raymundo Cassani '' [[https://en.wikipedia.org/wiki/Corticomuscular_coherence|Corticomuscular coherence]] relates to the synchrony between electrophysiological signals (MEG, EEG or ECoG) recorded from the contralateral motor cortex, and EMG signal from a muscle during voluntary movement. This synchrony has its origin mainly in the descending communication in corticospinal pathways between primary motor cortex (M1) and muscles. This tutorial replicates the processing pipeline and analysis presented in the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|Analysis of corticomuscular coherence]] FieldTrip tutorial. |
''Authors: Raymundo Cassani, Francois Tadel & Sylvain Baillet.'' [[https://en.wikipedia.org/wiki/Corticomuscular_coherence|Corticomuscular coherence]] measures a degree of similarity between electrophysiological signals (MEG, EEG, ECoG sensor traces or source time series, especially over the contralateral motor cortex) and the EMG signal recorded from muscle activity during voluntary movement. This signal similarity is due mainly to the descending communication along corticospinal pathways between primary motor cortex (M1) and muscles. For consistency and reproducibility purposes across major software toolkits, the present tutorial replicates the processing pipeline "[[https://www.fieldtriptoolbox.org/tutorial/coherence/|Analysis of corticomuscular coherence]]" by FieldTrip. |
| Line 13: | Line 13: |
| [[Tutorials/Connectivity#Coherence|Coherence]] is a classic method to measure the linear relationship between two signals in the frequency domain. Previous studies ([[https://dx.doi.org/10.1113/jphysiol.1995.sp021104|Conway et al., 1995]], [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Kilner et al., 2000]]) have used coherence to study the relationship between MEG signals from M1 and muscles, and they have shown synchronized activity in the 15–30 Hz range during maintained voluntary contractions. | [[Tutorials/Connectivity#Coherence|Coherence]] measures the linear relationship between two signals in the frequency domain. Previous studies ([[https://dx.doi.org/10.1113/jphysiol.1995.sp021104|Conway et al., 1995]], [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Kilner et al., 2000]]) have reported cortico-muscular coherence effects in the 15–30 Hz range during maintained voluntary contractions. |
| Line 18: | Line 18: |
| The dataset is comprised of MEG (151-channel CTF MEG system) and bipolar EMG (from left and right extensor carpi radialis longus muscles) recordings from one subject during an experiment in which the subject had to lift her hand and exert a constant force against a lever. The force was monitored by strain gauges on the lever. The subject performed two blocks of 25 trials in which either the left or the right wrist was extended for about 10 seconds. In addition to the MEG and EMG signals, EOG signal was recorded to assist the removal of ocular artifacts. Only data for the left wrist will be analyzed in this tutorial. | The dataset comprises recordings from MEG (151-channel CTF MEG system) and bipolar EMG (from left and right extensor carpi radialis longus muscles) from one participant who was tasked to lift their hand and exert a constant force against a lever for about 10 seconds. The force was monitored by strain gauges on the lever. The participant performed two blocks of 25 trials using either the left or right wrist. EOG signals were also recorded, which will be useful for detection and attenuation of ocular artifacts. We will analyze the data from the left-wrist trials in the present tutorial. Replicating the pipeline with right-wrist data is a good exercise to do next! |
| Line 21: | Line 21: |
| * '''Requirements''': You should have already followed all the [[Tutorials|get-started tutorials]] and you have a working copy of Brainstorm installed on your computer. | * '''Requirements''': Please make sure you have completed the [[Tutorials|get-started tutorials]] and that you have a working copy of Brainstorm installed on your computer. |
| Line 23: | Line 23: |
| * Download the `SubjectCMC.zip` file from FieldTrip FTP server:<<BR>> ftp://ftp.fieldtriptoolbox.org/pub/fieldtrip/tutorial/SubjectCMC.zip * Unzip it in a folder that is not in any of the Brainstorm folders (program or database folder). |
* Download `SubjectCMC.zip` from FieldTrip's FTP server:<<BR>> ftp://ftp.fieldtriptoolbox.org/pub/fieldtrip/tutorial/SubjectCMC.zip * Unzip the .zip in a folder not located in any of current Brainstorm's folders (the app per se or its database folder). |
| Line 26: | Line 26: |
| * Start Brainstorm (Matlab scripts or stand-alone version). | * Launch Brainstorm (via Matlab's command line or use Brainstorm's Matlab-free stand-alone version). |
| Line 29: | Line 29: |
| The next sections describe: importing the subject's anatomy, reviewing raw data, managing event markers, pre-processing the EMG and MEG signals, epoching and importing recordings, and computing coherence in the sensor and source level. | The next sections describe how to import the participant's anatomical data, review raw data, manage event markers, pre-process EMG and MEG signals, epoch and import recordings for further analyzes, with a focus on computing coherence at the sensor (scalp) and brain map (sources) levels. |
| Line 32: | Line 32: |
| * Right-click on the '''TutorialCMC''' node then '''''New subject > Subject01'''''.<<BR>>Keep the default options you defined for the protocol. * Switch to the '''Anatomy''' view of the protocol. |
* Right-click on the newly created '''TutorialCMC''' node in your Brainstorm data tree then '''''New subject > Subject01'''''.<<BR>>Keep the default options defined for the study (aka "protocol" in Brainstorm's jargon). * Switch to the '''Anatomy''' view of the study. |
| Line 36: | Line 36: |
| * Set the file format: '''All MRI file (subject space)''' | * Select the adequate file format from the pull-down menu: '''All MRI file (subject space)''' |
| Line 39: | Line 39: |
| * Compute MNI normalization, in the '''MRI viewer''' click on '''Click here to compute MNI normalization''', use the '''maff8''' method. When the normalization is complete, verify the correct location of the fiducials and click on '''Save'''. | * Register the individual anatomy to MNI brain space, for standardization of coordinates: in the '''MRI viewer''' click on '''Click here to compute MNI normalization''', use the '''maff8''' method. When the normalization is complete, verify that the locations of the anatomical fiducials are adequate (essentially that they are indeed near the left/right ears and right above the nose) and click on '''Save'''. |
| Line 46: | Line 46: |
| Once the MRI has been imported and normalized, we need to segment the head and brain tissues to obtain the surfaces that are needed to generate a realistic [[Tutorials/HeadModel|head model (or forward model)]]. | We then need to segment the head tissues to obtain the surfaces required to derive a realistic MEG [[Tutorials/HeadModel|head model (aka "forward model")]]. |
| Line 53: | Line 53: |
| * Rick-click on the '''tissues''' node and select '''''Generate triangular meshes''''' * Select the 5 layers to mesh |
* Rick-click on the '''tissues''' node and select '''''Generate triangular meshes.''''' * Select the 5 layers to mesh. |
| Line 60: | Line 60: |
| As output, there is a set of (head and brain) surface files that will be used for the head model computation. | A set of (head and brain) surface files are now available for further head modelling (see below). |
| Line 64: | Line 64: |
| By displaying the surfaces, we can note that the '''cortex''', which is related to the gray matter (shown in red) overlaps heavily with the '''innerskull''' surface (shown in gray), so it cannot be used to compute a [[Tutorials/TutBem|BEM forward model using OpenMEEG]]. However, as we are dealing with MEG signals, we can still compute the forward model with the [[Tutorials/HeadModel#Forward_model|overlapping-spheres method]], and obtain similar results. We can also notice that the '''cortex''' and '''white''' surfaces obtained with the method above do not register accurately the cortical surface, they can be used for [[Tutorials/TutVolSource|volume-based source estimation]], which is based on a volume grid of source points; but they may not be used for surface-based source estimation. Better surfaces can be obtained by doing MRI segmentation with [[Tutorials/SegCAT12|CAT12]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. | You can display the surfaces by double-clicking on these new nodes. There are a couple of issues with the structural data available from this tutorial. Note how the '''cortex''' (shown in red) overlaps with the '''innerskull''' surface (shown in gray). For this reason, the [[Tutorials/TutBem|BEM forward model cannot be derived with OpenMEEG]]. We will use an analytical approximationusing the [[Tutorials/HeadModel#Forward_model|overlapping-spheres method]], which in MEG has been shown to be adequately accurate for most studies. Note also how the '''cortex''' and '''white''' surfaces obtained do not register accurately with the cortical surface. We will therefore use a [[Tutorials/TutVolSource|volume-based source estimation]] approach based on a volumic grid of elementary MEG source across the cerebrum (not a surface-constrained source model). We encourage users to [[Tutorials/SegCAT12|CAT12]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]] to obtain surface segmentations of higher quality. |
| Line 70: | Line 70: |
| * Right-click on Subject01 then go to the menu '''''Add MNI parcellation > AAL3 '''''. The menu will appear as '''''Download: AAL3 ''''' if the atlas is not in your system. == Access the recordings == === Link the recordings === * Switch to the '''Functional data''' view (X button). |
* Right-click on Subject01 then go to the menu '''''Add MNI parcellation > AAL3'''''. The menu will appear as '''''Download: AAL3''''' if the atlas is not in your system. Once the MNI atlas is downloaded, an atlas node (ICON) appears in the database explorer and the atlas is displayed in the the MRI viewer. . {{attachment:gui_mni_aal3.png}} <<BR>> == Review the MEG and EMG recordings == === Link the recordings to Brainstorm's database === * Switch now to the '''Functional data''' view (X button). |
| Line 76: | Line 78: |
| * Select the file format: '''MEG/EEG: CTF(*.ds; *.meg4; *.res4)''' | * Select the file format of current data from the pulldown menu options: '''MEG/EEG: CTF(*.ds; *.meg4; *.res4)''' |
| Line 79: | Line 81: |
| A new folder and its content are now visible in the database explorer: * The '''SubjectCMC''' folder represents the MEG dataset linked to the database. Note the tag "RAW" in the icon of the folder, this means that the files are considered as raw continuous files. * The '''CTF channels (191)''' node is the '''channel file''' and defines the types and names of channels that were recorded, the position of the sensors, the head shape and other various details. This information has been read from the MEG datasets and saved as a new file in the database. The total number of data channels recorded in the file is indicated between parenthesis '''(191)'''. * The '''Link to raw file''' node is a '''link to the original file''' that was selected. All the relevant metadata was read from the MEG dataset and copied inside the link itself (sampling rate, number of samples, event markers and other details about the acquisition session). As it is a link, no MEG recordings were copied to the database. When we open this file, the values are read directly from the original files in the .ds folder. [[Tutorials/ChannelFile#Review_vs_Import|More information]]. |
A new folder is now created in Brainstorm's database explorer and contains: * '''SubjectCMC''': a folder that provides access to the MEG dataset. Note the "RAW" tag over the icon of the folder, indicating the files contain unprocessed, continuous data. * '''CTF channels (191)''': a node containing '''channel information''' with all channel types, names locations, etc. The number of channels available (MEG, EMG, EOG etc.) is indicated between parentheses''' (here, 191'''). * '''Link to raw file''' provides access to '''to the original data file'''. All the relevant metadata was read from the dataset and copied inside the node itself (e.g., sampling rate, number of time samples, event markers). Note that Brainstorm's logic is not to import/duplicate the raw unprocessed data directly into the database. Instead, Brainstorm provides a link to that raw file for further review and data extraction ([[Tutorials/ChannelFile#Review_vs_Import|more information]]). |
| Line 88: | Line 90: |
| * Right-click on the '''CTF channels (191)''' node, then '''''Display sensors > CTF helmet''''' and '''''Display sensors > MEG ''''' to show a surface that represents the inner surface the helmet, and the MEG sensors respectively. Try [[Tutorials/ChannelFile#Display_the_sensors|additional display menus]]. | * Right-click on the '''CTF channels (191)''' node, then select '''''Display sensors > CTF helmet''''' from the contextual menu and '''''Display sensors > MEG. '''''This will open a new display window showing the inner surface of the MEG helmet, and the lo MEG sensors respectively. Try [[Tutorials/ChannelFile#Display_the_sensors|additional display menus]]. |
| Line 93: | Line 95: |
| * Right-click on the '''Link to raw file''' node, then '''''Switch epoched/continuous''''' to convert the file to '''continuous'''. * Right-click on the '''Link to raw file''' node, then '''''MEG > Display time series''''' (or double-click on the node). This opens a new time series figure and enable the '''''Time''''' panel and the '''''Record''''' tab in the main Brainstorm window. Controls in this panel and tab are used to [[Tutorials/ReviewRaw|explore the time series]]. * In addition we can display the EMG signals, right-click on the '''Link to raw file''' node, then '''''EMG > Display time series'''''. |
* Right-click on the '''Link to raw file''' node, then '''''Switch epoched/continuous''''' to convert the file to '''continuous''', a technical detail proper to CTF file formatting. * Right-click again on the '''Link to raw file''' node, then '''''MEG > Display time series''''' (or double-click on the node). This will open a new visualization window to explore data time series, also enabling the '''''Time''''' panel and the '''''Record''''' tab in the main Brainstorm window (see how to best use all controls in this panel and tab to [[Tutorials/ReviewRaw|explore data time series]]). * We will also display EMG traces by right-clicking on the '''Link to raw file''' node, then '''''EMG > Display time series'''''. |
| Line 100: | Line 102: |
| The colored dots on top of the recordings in the time series figures represent the [[Tutorials/EventMarkers|event markers]] (or triggers) saved in this dataset. In addition to these events, the start of the either left or right trials is saved in the auxiliary channel named '''Stim'''. To add these markers: * With the time series figure open, in the '''''Record''''' tab go to '''''File > Read events from channel'''''. Now, in the options for the '''Read from channel''' process, set '''Event channels''' = `Stim`, select '''Value''', and click '''Run'''. |
The colored dots above the data time series indicate [[Tutorials/EventMarkers|event markers]] (or triggers) saved with this dataset. The trial onset information of the left-wrist and right-wrist trials is saved in an auxiliary channel of the raw data named '''Stim'''. To add these markers, these events need to be decoded as follows: * While the time series figure is open, go to the '''''Record''''' tab and '''''File > Read events from channel'''''. From the options of the '''Read from channel''' process window, set '''Event channels''' = `Stim`, select '''Value''', and click '''Run'''. |
| Line 106: | Line 108: |
| This creates new events shown in the '''''Events''''' section of the tab. We are only interested in the events from '''U1''' to '''U25''' that correspond to the 25 left trials. Thus we will delete the other events, and merge the left trial events. * Delete all the other events: select the events to delete with '''Ctrl+click''', when done go the menu '''''Events > Delete group''''' and confirm. Alternatively, you can do '''Ctrl+A''' to select all the events and then deselect the '''U1''' to '''U25''' events. * To be in line with the original FieldTrip tutorial, we will reject the trial 7. Select the events '''U1''' to '''U6''' and '''U8''' to '''U25''' events, then go the menu '''Events > Merge group''' and enter the label '''Left'''. |
This procedure creates new event markers now shown in the '''''Events''''' section of the tab. along with previous event categories. In this tutorial, we will only use events '''U1''' through '''U25''', which correspond to how each of the 25 left-wrist trials had been encoded in the study. We will now delete other events of no interest, and merge the left trial events under a single event category, for convenience. * Delete other events: select the events to delete in the event box/list with '''Ctrl+click''', then in the menu '''''Events > Delete group''''' and confirm. Alternatively, you can selected all events with '''Ctrl+A''' and deselect the '''U1''' to '''U25''' events by clicking on them. * To make sure we reproduce FieldTrip's tutorial, we need to reject trial #7: Select events '''U1''' to '''U6''' and '''U8''' to '''U25''', then from the '''Events '''menu, select''' Merge group''' and type ina new label ('''Left''') to describe this is the left-wrist condition. |
| Line 113: | Line 115: |
| These events are located at the beginning of the 10 s trials of left wrist movement. In the sections below, we will compute the coherence for 1 s epochs for the first 8 s of the trial, thus we need to create extra events. | These events correspond to the beginning of 10-s trials of left-wrist movements. We will compute coherence over 1-s epochs over the first 8 s of each trial. To that purpose, we will now create extra events to define these epochs. |
| Line 116: | Line 118: |
| * For each copy of the '''Left''' events, we need to add a time offset of 1 s for '''Left02''', 2 s for '''Left03''', and so on. Select the event group to add the offset, then go to the menu '''''Events > Delete group'''''. | * For each copy of the '''Left''' events, we will add a time offset of 1 s for '''Left02''', 2 s for '''Left03''', and so on. Select the '''Left '''event group to add a 1,000 ms time offset, by going to the menu '''''Events > Add time offset'', '''enter 1,000 in the text box. Repeat for each other group, entering 2,000, then 3,000 etc. |
| Line 120: | Line 122: |
| * Finally, merge all the '''Left*''' events into '''Left''', and select '''''Save modifications''''' in the '''''File''''' menu in the '''''Record''''' tab. | * Once done for '''Left_08''', merge all these '''Left*''' events into a single '''Left '''category, and select '''''Save modifications''''' in the '''''File''''' menu in the '''''Record''''' tab. |
| Line 209: | Line 211: |
| In the case of stereotypical artifacts, as it is the case of the eye blinks and heartbeats, it is possible to identify their characteristic spatial distribtuion, and then remove it from MEG signals with methods such as Signal-Space Projection (SSP). For more details, consult the tutorials on [[Tutorials/ArtifactsDetect|detection]] and [[Tutorials/ArtifactsSsp|removal of artifacts with SSP]]. The dataset of this tutorial contains an EOG channel but not ECG signal, thus will perform only removal of eye blinks. | In the case of stereotypical artifacts, as it is the case of the eye blinks and heartbeats, it is possible to identify their characteristic spatial distribution, and then remove it from MEG signals with methods such as Signal-Space Projection (SSP). For more details, consult the tutorials on [[Tutorials/ArtifactsDetect|detection]] and [[Tutorials/ArtifactsSsp|removal of artifacts with SSP]]. The dataset of this tutorial contains an EOG channel but not ECG signal, thus will perform only removal of eye blinks. |
| Line 224: | Line 226: |
| * To [[Tutorials/ArtifactsSsp|remove blink artifacts with SSP]] go to '''''Artifacts > SSP: Eyeblinks''''', and use the parameters: | * To [[Tutorials/ArtifactsSsp|remove blink artifacts with SSP]] go to '''''Artifacts > SSP: Eye blinks''''', and use the parameters: |
| Line 324: | Line 326: |
| Before estimating the brain source, we need to compute the '''head model''' and the '''noise covariance'''. | In this tutorial we will perform source modelling using the [[Tutorials/HeadModel#Dipole_fitting_vs_distributed_models|distributed model]] approach for two sources spaces: '''cortex surface''' and '''MRI volume'''. In the first one the location of the sources is constrained to the cortical surface obtained when the subject anatomy was imported. For the second source space, the sources are uniformly distributed in the entire brain volume. Before estimating the brain sources, we need to compute '''head model''' and the '''noise covariance'''. Note that a head model is required for each source space. |
| Line 327: | Line 329: |
| The head model describes how neural electric currents produce magnetic fields and differences in electrical potentials at external sensors, given the different head tissues. This model is independent of sensor recordings. See the [[Tutorials/HeadModel|head model tutorial]] for more details. | The head model describes how neural electric currents produce magnetic fields and differences in electrical potentials at external sensors, given the different head tissues. This model is independent of sensor recordings. See the [[Tutorials/HeadModel|head model tutorial]] for more details. Each source space, requires its own head model. ==== Cortex surface ==== |
| Line 330: | Line 333: |
| * '''Source space''' = `Cortex` | * '''Comment''' = `Overlapping spheres (surface)` * '''Source space''' = `Cortex surface` |
| Line 333: | Line 337: |
| As result will have the '''Overlapping spheres''' head model in the database explorer. . {{attachment:pro_head_model.png||width="90%"}} |
Keep in mind that the number of sources (vertices) in this head model is '''10,000''', and was defined when when the subject anatomy was imported. . {{attachment:pro_head_model_srf.png||width="40%"}} The (ICON) '''Overlapping spheres (surface)''' head model will appear in the database explorer. ==== MRI volume ==== * In the '''SubjectCMC_clean_notch_high_abs''', right-click the '''CTF channels (191)''' node and select '''''Compute head model'''''. Keep the default options: * '''Comment''' = `Overlapping spheres (volume)` * '''Source space''' = `MRI volume` * '''Forward model''' = `Overlapping spheres`. . {{attachment:pro_head_model_vol.png||width="40%"}} * The '''Volume source grid''' window pop-up, to define the volume grid. Use the following parameters, that will lead to an estimated number of '''12,200''' grid points. * Select '''Regular grid''' and '''Brain''' * '''Grid resolution''' = `5 mm` . {{attachment:pro_grid_vol.png||width="50%"}} The '''Overlapping spheres (volume)''' node will be added to the database explorer. The green color indicates the default head model for the folder. . {{attachment:tre_head_models.png||width="50%"}} |
| Line 338: | Line 362: |
| For MEG it is [[Tutorials/NoiseCovariance#The_case_of_MEG|recommended]] to derive the noise covariance from empty room recordings. However, as we do not have those recordings in the dataset, we can compute the noise covariance from the MEG signals before the trials. See the [[Tutorials/NoiseCovariance|noise covariance tutorial]] for more details. * In the raw '''SubjectCMC_clean_notch_high_abs''', right click the '''Raw | clean | notch(...''' node and select '''''Noise covariance > Compute from recordings'''''. As parameters select a '''Baseline''' from `18 - 30 s`, and the '''Block by block''' option. . {{attachment:pro_noise_cov.png||width="40%"}} * Lastly, we need to copy the '''Noise covariance''' node to the '''SubjectCMC_clean_notch_high_abs''' folder with the head model. This can be done with the shortcuts `Ctrl-C` and `Ctrl-V`. |
For MEG recordings it is [[Tutorials/NoiseCovariance#The_case_of_MEG|recommended]] to derive the noise covariance from empty room recordings. However, as we do not have those recordings in the dataset, we can compute the noise covariance from the MEG signals before the trials. See the [[Tutorials/NoiseCovariance|noise covariance tutorial]] for more details. * In the raw '''SubjectCMC_clean_notch_high_abs''', right-click the '''Raw | clean | notch(...''' node and select '''''Noise covariance > Compute from recordings'''''. As parameters select: * '''Baseline''' from `18 - 30 s` * Select the '''Block by block''' option. . {{attachment:pro_noise_cov.png||width="60%"}} * Lastly, copy the '''Noise covariance''' node to the '''SubjectCMC_clean_notch_high_abs''' folder with the head model. This can be done with the shortcuts `Ctrl-C` and `Ctrl-V`. . {{attachment:tre_covmat.png||width="40%"}} |
| Line 347: | Line 375: |
| With the help of the '''head model''' and the '''noise covariance''', we can solve the '''inverse problem''' by computing an '''inverse kernel''' that will estimate the brain activity that gives origin to the observed recordings in the sensors. See the [[Tutorials/SourceEstimation|source estimation tutorial]] for more details. * To compute the inversion kernel, right click in the '''Overlapping spheres''' head model and select '''Compute sources [2018]'''. With the parameters: '''Minimum norm imaging''', '''dSPM''' and '''Constrained'''. . {{attachment:pro_sources.png||width="40%"}} The inversion kernel '''dSPM-unscaled: MEG(Constr) 2018''' was created, and note that the each recordings node has an associated source link. . {{attachment:gui_inv_kernel.png||width="90%"}} |
Noe that the '''head model(s)''' and '''noise covariance''' have been computed, we can use the [[Tutorials/SourceEstimation#Method|minimum norm imaging]] method to solve the '''inverse problem'''. The result is a linear '''inversion kernel''', that estimates the source brain activity that gives origin to the observed recordings in the sensors. Note that, an inversion kernel is obtained for each of the head models: '''surface''' and '''volume'''. See the [[Tutorials/SourceEstimation|source estimation tutorial]] for more details. ==== Cortex surface ==== * Compute the inversion kernel, right-click in the '''Overlapping spheres (surface)''' head model and select '''Compute sources [2018]'''. With the parameters: * '''Minimum norm imaging''' * '''Current density map''' * '''Unconstrained''' * '''Comment''' = `MN: MEG (surface)` . {{attachment:pro_sources_srf.png||width="40%"}} The inversion kernel (ICON) '''MN: MEG (surface)(Unconstr) 2018''' is created, and added to the database explorer. ==== MRI volume ==== * Compute the inversion kernel, right-click in the '''Overlapping spheres (volume)''' head model and select '''Compute sources [2018]'''. With the parameters: * '''Minimum norm imaging''' * '''Current density map''' * '''Unconstrained''' * '''Comment''' = `MN: MEG (volume)` . {{attachment:pro_sources_vol.png||width="40%"}} The inversion kernel (ICON) '''MN: MEG (volume)(Unconstr) 2018''' is created, and added to the database explorer. The green color in the name indicates the current default head model. In addition, note that each trial has now '''two''' associated source link (ICON) nodes. One obtained with the '''MN: MEG (surface)(Unconstr) 2018''' kernel and the other obtained with the '''MN: MEG (volume)(Unconstr) 2018''' kernel. . {{attachment:gui_inverse_kernel.png||width="60%"}} === Scouts === From the [[#head_model|head model section]], we notice that the '''cortex''' and '''volume''' grid have around '''10,000''' vertices each, thus as many sources were estimated. As such, it is not practical to compute coherence between the left EMG signal and the signal of each source. A way to address this issue is with the use of regions of interest also known as '''scouts'''. Thus, there are [[Tutorials/Scouts|surface scouts]] and [[Tutorials/TutVolSource#Volume_scouts|volume scouts]]. Let's define scouts for the different source spaces. ==== Surface scouts ==== * In the source link '''MN: MEG (surface)(Unconstr) 2018''' node for one of the trials, right-click and select '''''Cortical activations > Display on cortex'''''. In the '''''Surface''''' tab, set the '''''Amplitude''''' slider to `100%` to hide all the sources. . {{attachment:fig_sources_srf.png||width="100%"}} * In the '''''Scout''''' tab, select the menu '''''Atlas > From subject anatomy > AAL3 (MNI-linear)'''''. This will create the '''From volume: AAL3''' set of surface scouts. By clicking in the different scouts, at the bottom of the list, the number of vertices it contains and the approximate area in cm2 is shown. Activate the (ICON) '''Show only the selected scouts''' option to narrow down the shown scouts. . {{attachment:fig_scouts_srf.png||width="90%"}} {{{#!wiki caution [TODO] A note, that the definition of scouts is far from perfect, but can give us a good idea of the surface projections of the MNI parcellations (described in the importing anatomy section). }}} * Close the figure. ==== Volume scouts ==== * In the source link '''MN: MEG (volume)(Unconstr) 2018''' node for one of the trials, right-click and select '''''Cortical activations > Display on MRI (3D): Subject CMC'''''. In the '''''Surface''''' tab, set the '''''Amplitude''''' slider to `100%` to hide all the sources. . {{attachment:fig_sources_vol.png||width="100%"}} * In the '''''Scout''''' tab, select the menu '''''Atlas > From subject anatomy > AAL3 (MNI-linear)'''''. This will create the '''Volume 12203: AAL3''' set of volume scouts. By clicking in the different scouts, at the bottom of the list, the number of vertices it contains and the approximate volume in cm3 is shown. Activate the (ICON) '''Show only the selected scouts''' option to narrow down the shown scouts. . {{attachment:fig_scouts_vol.png||width="90%"}} * Close the figure. |
| Line 358: | Line 430: |
| {{{#!wiki warning '''[TO DISCUSS among authors]''' Better source localization can be obtained by performing MRI segmentation with CAT12. Although it adds between ~45min of additional processing. We may want to provide the already processed MRI. Thoughts? }}} From the earlier section [[#Importing_anatomy_data|importing anatomy data]], we can observe that the cortex surface has '''10,000''' vertices, thus as many sources were estimated. AS it can be seen, it is not practical to compute coherence between the left EMG signal and each source. A way to address this issue is with the use of regions of interest or [[Tutorials/Scouts|Scouts]]. * Double-click the source link for any of the trials to visualize the source space. This will enable the tab '''Scout''' in the main Brainstorm window. . {{attachment:scout_tab.png||width="70%"}} * In the '''Scout''' tab select the menu '''Atlas > Surface clustering > Homogeneous parcellation (deterministic)''', and set the '''Number of scouts''' to `80`. . {{attachment:parcellation.png||width="70%"}} It is important to note that the coherence will be performed between a sensor signal (EMG) and source signals in the scouts. |
Coherence in the source level is computed between a sensor signal (EMG) and source signals in the (surface or volume) scouts. === Coherence with surface scouts === To facilitate the selection of the indicated files to compute this coherence, let's [[Tutorials/PipelineEditor#Search_Database|search in the database]] the recordings and the source link files obtained wit the '''MN: MEG (surface)(Unconstr) 2018''' kernel. * Click on the magnifying glass (ICON) above the database explorer to open up the search dialog, and select '''''New search'''''. * Set the search query to look for files that '''are named (surface)''' or '''are named Left'''. This is done with the following configuration. . {{attachment:gui_search_srf.png||width="70%"}} By performing the search, a new tab called '''''(surface)''''' appears in the database explorer. This new tab contains the recordings and ONLY the source link for the surface space. . {{attachment:tre_search_srf.png||width="40%"}} * Change to the '''Process2''' tab, and drag-and-drop the '''Left (192 files)''' trial group into the '''Files A''' and into the '''Files B''' boxes. And select '''Process recordings''' for Files A, and '''Process sources''' for Files B. Note that blue labels over the '''Files A''' and the '''Files B''' boxes indicate that there are 185 files per box. . {{attachment:process2.png||width="80%"}} Open the '''Pipeline editor''' and add two process: * Add the process '''Connectivity > Coherence AxB [2021]''' with the following parameters: * '''Time window''' = `0 - 1000 ms` or check '''All file''' * '''Source channel (A)''' = `EMGlft` * Check '''Use scouts (B)''' * Select `From volume: AAL3` in the drop-down list (these are surface scouts) * Select all the scouts (shortcut `Ctrl-A`) * '''Scout function''' = `Mean` * '''When to apply''' = `Before` * Do not '''Remove evoked responses from each trial''' * '''Magnitude squared coherence''', '''Window length''' = `0.5 s` * '''Overlap''' = `50%` * '''Highest frequency''' = `80 Hz` * '''Average cross-spectra'''. * Add the process '''File > Add tag''' with the following parameters: * '''Tag to add''' = `(surface)` * Select '''Add to file name''' * Run the pipeline . {{attachment:pro_coh_srf.png||width="100%"}} * Double-click on the resulting node '''mscohere(0.6Hz,555win): Left (#1) | (surface)''' to display the coherence spectra. Also open the result node as image with '''Display image''' in its context menu. * To verify the location of the scouts on the cortex surface, double-click one of the (surface) source link for any of the trials. In the '''Surface''' tab, set the '''Amplitude''' threshold to `100%` to hide all the cortical activations. Lastly, in the '''Scouts''' tab, select the `From volume: AAL3` atlas in the drop-down list, select '''Show only the selected scouts''' and the '''Show/hide the scout labels'''. Note that the plots are linked by the scout selected in the '''image''' representation of the coherence results. . {{attachment:res_coh_srf.png||width="100%"}} From the results we can see that the peak at 14.65 Hz corresponds to the '''Precentral R''' scout, which encompasses the right primary motor cortex, as expected. These results are inline with the ones in the literature. === Coherence with volume scouts === Similar to coherence with surface scouts, a search is needed to select the recordings and the source link files obtained wit the '''MN: MEG (volume)(Unconstr) 2018''' kernel. * Click on the magnifying glass (ICON) above the database explorer to open up the search dialog, and select '''''New search'''''. Set the search query to look for files that '''are named (volume)''' or '''are named Left'''. This is done with the following configuration. . {{attachment:gui_search_vol.png||width="70%"}} . {{attachment:tre_search_vol.png||width="40%"}} |
| Line 377: | Line 494: |
| * Run the process '''Connectivity > Coherence AxB [2021]''' with the following parameters: * '''All file''', '''Source channel (A)''' = `EMGlft`, check '''Use scouts (B)''' * Select `Surface clustering: 80` in the drop-down list, and select all the scouts (shortcut `Ctrl-A`). * '''Scout function''' = `Mean`, and '''When to apply''' = `Before`. Do not '''Remove evoked responses from each trial'''. * '''Magnitude squared coherence''', '''Window length''' = `0.5 s`, '''Overlap''' = `50%`, '''Highest frequency''' = `80 Hz`, and '''Average cross-spectra'''. . {{attachment:pro_coh_ab.png||width="40%"}} === Results with FieldTrip MRI segmentation === * Double-click on the resulting node '''mscohere(0.6Hz,555win): Left (#1)''' to display the coherence spectra. Also open the result node as image with '''Display image''' in its context menu. * To verify the location of the scouts on the cortex surface, double-click the source link for any of the trials. In the '''Surface''' tab, set the '''Amplitude''' threshold to `100%` to hide all the cortical activations. Laslty, in the '''Scouts''' tab, select the '''Show only the selected scouts''' and the '''Show/hide the scout labels'''. Note that the plots are linked by the scout selected in the '''image''' representation of the coherence results. . {{attachment:res_coh_ab_ft.png||width="100%"}} === Results with FieldTrip MRI segmentation === {{{#!wiki warning '''[TO DISCUSS among authors]''' Same as the previous section but using the surface from CAT, and using DK atlas. {{attachment:res_coh_ab_cat.png||width="100%"}} }}} {{{#!wiki warning '''[TO DISCUSS among authors]''' In addition I barely ran the Coherence (as it took up to 30GB) for all the vertices vs EMG Left for the source estimation using the FielfTrip and the CAT12 segmentations |
Open the '''Pipeline editor''' and add two process: * Add the process '''Connectivity > Coherence AxB [2021]''' with the following parameters: * '''Time window''' = `0 - 1000 ms` or check '''All file''' * '''Source channel (A)''' = `EMGlft` * Check '''Use scouts (B)''' * Select `Volume 12203: AAL3` in the drop-down list (these are volume scouts) * Select all the scouts (shortcut `Ctrl-A`) * '''Scout function''' = `Mean` * '''When to apply''' = `Before` * Do not '''Remove evoked responses from each trial''' * '''Magnitude squared coherence''', '''Window length''' = `0.5 s` * '''Overlap''' = `50%` * '''Highest frequency''' = `80 Hz` * '''Average cross-spectra'''. * Add the process '''File > Add tag''' with the following parameters: * '''Tag to add''' = `(surface)` * Select '''Add to file name''' * Run the pipeline . {{attachment:pro_coh_vol.png||width="100%"}} * Double-click on the resulting node '''mscohere(0.6Hz,555win): Left (#1) | (volume)''' to display the coherence spectra. Also open the result node as image with '''Display image''' in its context menu. * To verify the location of the scouts on the cortex surface, open one of the (volume) source link for any of the trials with the as MRI (3D), in the context menu select '''''Display cortical activations > Display on MRI (3D): SubjectCMC '''''. In the '''Surface''' tab, set the '''Amplitude''' threshold to `100%` to hide all the cortical activations. Lastly, in the '''Scouts''' tab, select the `Volume 12203: AAL3` atlas in the drop-down list, select the '''Show only the selected scouts''' and the '''Show/hide the scout labels'''. Note that the plots are linked by the scout selected in the '''image''' representation of the coherence results. . {{attachment:res_coh_vol.png||width="100%"}} From the results we can see that the peak at 14.65 Hz corresponds to the '''Precentral R''' scout, which encompasses the right primary motor cortex, as expected. These results are inline with the ones in the literature. <<TAG(Advanced)>> == Coherence with all sources (no scouts) == {{{#!wiki caution * We could downsample the surface and create a more sparse volume grid . OR * Refactor the coherence process to accumulate the auto- and cross-spectra outside of the function . OR }}} === Comparison of cortex surface with FieldTrip and CAT === {{{#!wiki caution '''[TO DISCUSS among authors]''' This image and GIF are just for reference. They were obtained with all the surface sources using FieldTrip and CAT derived surfaces. |
| Line 411: | Line 550: |
| {{{#!wiki warning | {{{#!wiki caution |
| Line 427: | Line 566: |
| {{{#!wiki warning | {{{#!wiki caution |
Corticomuscular coherence (MEG)
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Raymundo Cassani, Francois Tadel & Sylvain Baillet.
Corticomuscular coherence measures a degree of similarity between electrophysiological signals (MEG, EEG, ECoG sensor traces or source time series, especially over the contralateral motor cortex) and the EMG signal recorded from muscle activity during voluntary movement. This signal similarity is due mainly to the descending communication along corticospinal pathways between primary motor cortex (M1) and muscles. For consistency and reproducibility purposes across major software toolkits, the present tutorial replicates the processing pipeline "Analysis of corticomuscular coherence" by FieldTrip.
Contents
Background
Coherence measures the linear relationship between two signals in the frequency domain. Previous studies (Conway et al., 1995, Kilner et al., 2000) have reported cortico-muscular coherence effects in the 15–30 Hz range during maintained voluntary contractions.
IMAGE OF EXPERIMENT, SIGNALS and COHERENCE
Dataset description
The dataset comprises recordings from MEG (151-channel CTF MEG system) and bipolar EMG (from left and right extensor carpi radialis longus muscles) from one participant who was tasked to lift their hand and exert a constant force against a lever for about 10 seconds. The force was monitored by strain gauges on the lever. The participant performed two blocks of 25 trials using either the left or right wrist. EOG signals were also recorded, which will be useful for detection and attenuation of ocular artifacts. We will analyze the data from the left-wrist trials in the present tutorial. Replicating the pipeline with right-wrist data is a good exercise to do next!
Download and installation
Requirements: Please make sure you have completed the get-started tutorials and that you have a working copy of Brainstorm installed on your computer.
Download the dataset:
Download SubjectCMC.zip from FieldTrip's FTP server:
ftp://ftp.fieldtriptoolbox.org/pub/fieldtrip/tutorial/SubjectCMC.zip- Unzip the .zip in a folder not located in any of current Brainstorm's folders (the app per se or its database folder).
Brainstorm:
- Launch Brainstorm (via Matlab's command line or use Brainstorm's Matlab-free stand-alone version).
Select the menu File > Create new protocol. Name it TutorialCMC and select the options:
No, use individual anatomy,
No, use one channel file per acquisition run.
The next sections describe how to import the participant's anatomical data, review raw data, manage event markers, pre-process EMG and MEG signals, epoch and import recordings for further analyzes, with a focus on computing coherence at the sensor (scalp) and brain map (sources) levels.
Importing anatomy data
Right-click on the newly created TutorialCMC node in your Brainstorm data tree then New subject > Subject01.
Keep the default options defined for the study (aka "protocol" in Brainstorm's jargon).Switch to the Anatomy view of the study.
Right-click on the Subject01 node then Import MRI:
Select the adequate file format from the pull-down menu: All MRI file (subject space)
Select the file: SubjectCMC/SubjectCMC.mri
Register the individual anatomy to MNI brain space, for standardization of coordinates: in the MRI viewer click on Click here to compute MNI normalization, use the maff8 method. When the normalization is complete, verify that the locations of the anatomical fiducials are adequate (essentially that they are indeed near the left/right ears and right above the nose) and click on Save.
We then need to segment the head tissues to obtain the surfaces required to derive a realistic MEG head model (aka "forward model").
Right-click on the SubjectCMC MRI node, then MRI segmentation > FieldTrip: Tissues, BEM surfaces.
Select all the tissues (scalp, skull, csf, gray and white).
Click OK.
For the option Generate surface meshes select No.
After the segmentation is complete, a tissues node will be shown in the tree.
Rick-click on the tissues node and select Generate triangular meshes.
- Select the 5 layers to mesh.
- Use the default parameters:
number of vertices: 10,000
erode factor: 0
fill holes factor: 2
A set of (head and brain) surface files are now available for further head modelling (see below).
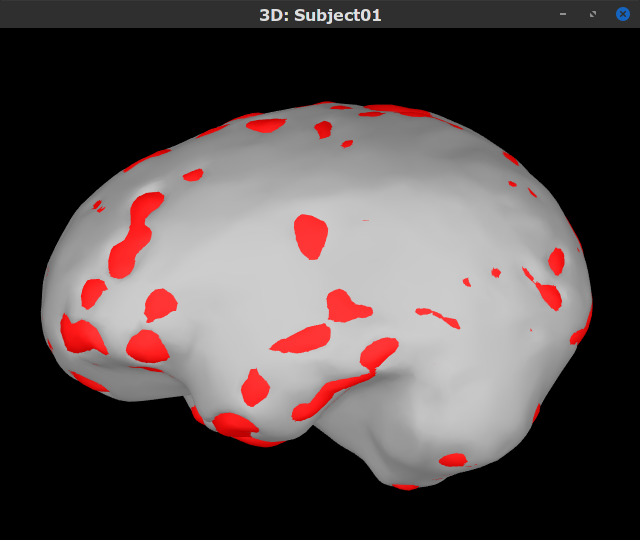
You can display the surfaces by double-clicking on these new nodes. There are a couple of issues with the structural data available from this tutorial. Note how the cortex (shown in red) overlaps with the innerskull surface (shown in gray). For this reason, the BEM forward model cannot be derived with OpenMEEG. We will use an analytical approximationusing the overlapping-spheres method, which in MEG has been shown to be adequately accurate for most studies. Note also how the cortex and white surfaces obtained do not register accurately with the cortical surface. We will therefore use a volume-based source estimation approach based on a volumic grid of elementary MEG source across the cerebrum (not a surface-constrained source model). We encourage users to CAT12 or FreeSurfer to obtain surface segmentations of higher quality.
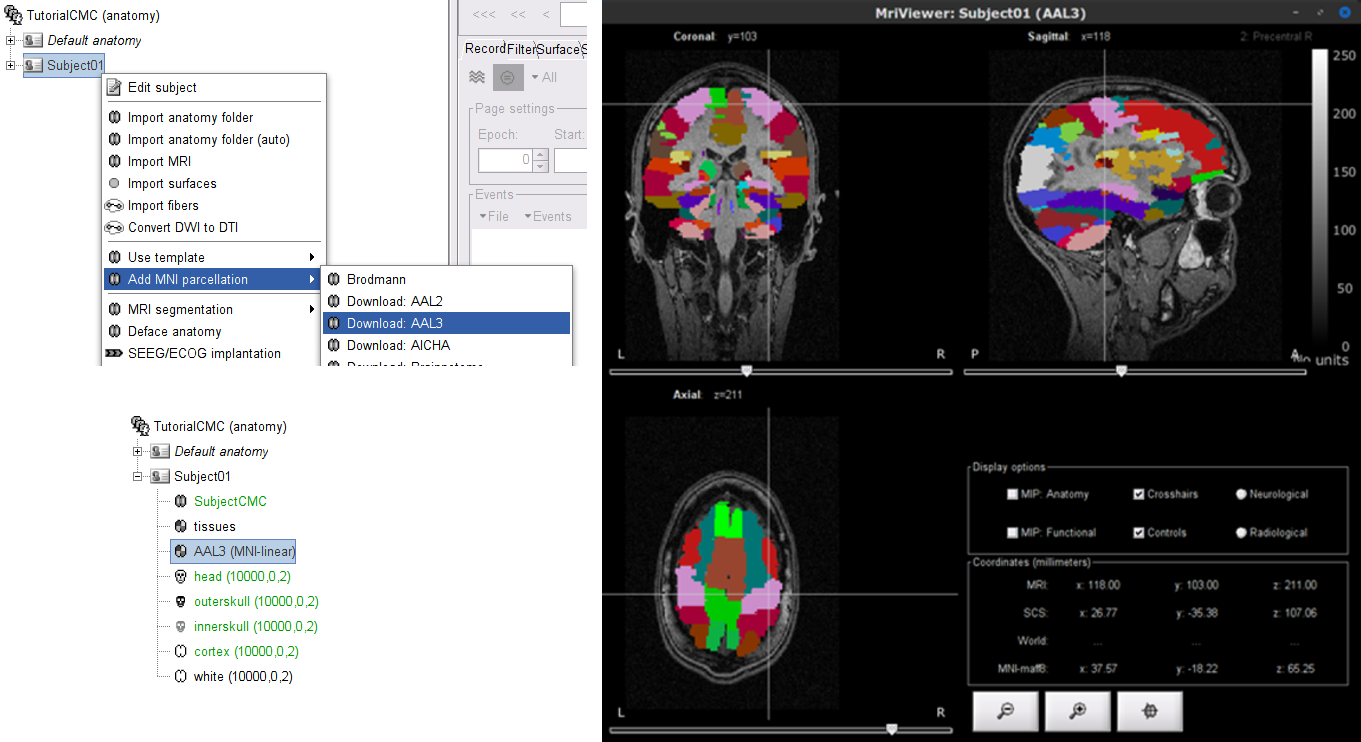
As the imported anatomy data is normalized in the MNI space, it is possible to apply use MNI parcellation templates to define anatomical regions of the brain of the subject. These anatomical regions can be used to create volume and surface scouts, which are convenient when performing the coherence analysis in the source level. Let's add the AAL3 parcellation to the imported data.
Right-click on Subject01 then go to the menu Add MNI parcellation > AAL3. The menu will appear as Download: AAL3 if the atlas is not in your system. Once the MNI atlas is downloaded, an atlas node (ICON) appears in the database explorer and the atlas is displayed in the the MRI viewer.
Review the MEG and EMG recordings
Link the recordings to Brainstorm's database
Switch now to the Functional data view (X button).
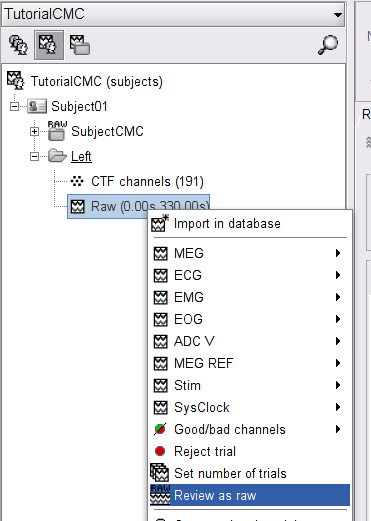
Right-click on the Subject01 node then Review raw file:
Select the file format of current data from the pulldown menu options: MEG/EEG: CTF(*.ds; *.meg4; *.res4)
Select the file: SubjectCMC.ds
A new folder is now created in Brainstorm's database explorer and contains:
SubjectCMC: a folder that provides access to the MEG dataset. Note the "RAW" tag over the icon of the folder, indicating the files contain unprocessed, continuous data.
CTF channels (191): a node containing channel information with all channel types, names locations, etc. The number of channels available (MEG, EMG, EOG etc.) is indicated between parentheses (here, 191).
Link to raw file provides access to to the original data file. All the relevant metadata was read from the dataset and copied inside the node itself (e.g., sampling rate, number of time samples, event markers). Note that Brainstorm's logic is not to import/duplicate the raw unprocessed data directly into the database. Instead, Brainstorm provides a link to that raw file for further review and data extraction (more information).
Display MEG helmet and sensors
Right-click on the CTF channels (191) node, then select Display sensors > CTF helmet from the contextual menu and Display sensors > MEG. This will open a new display window showing the inner surface of the MEG helmet, and the lo MEG sensors respectively. Try additional display menus.
Reviewing continuous recordings
Right-click on the Link to raw file node, then Switch epoched/continuous to convert the file to continuous, a technical detail proper to CTF file formatting.
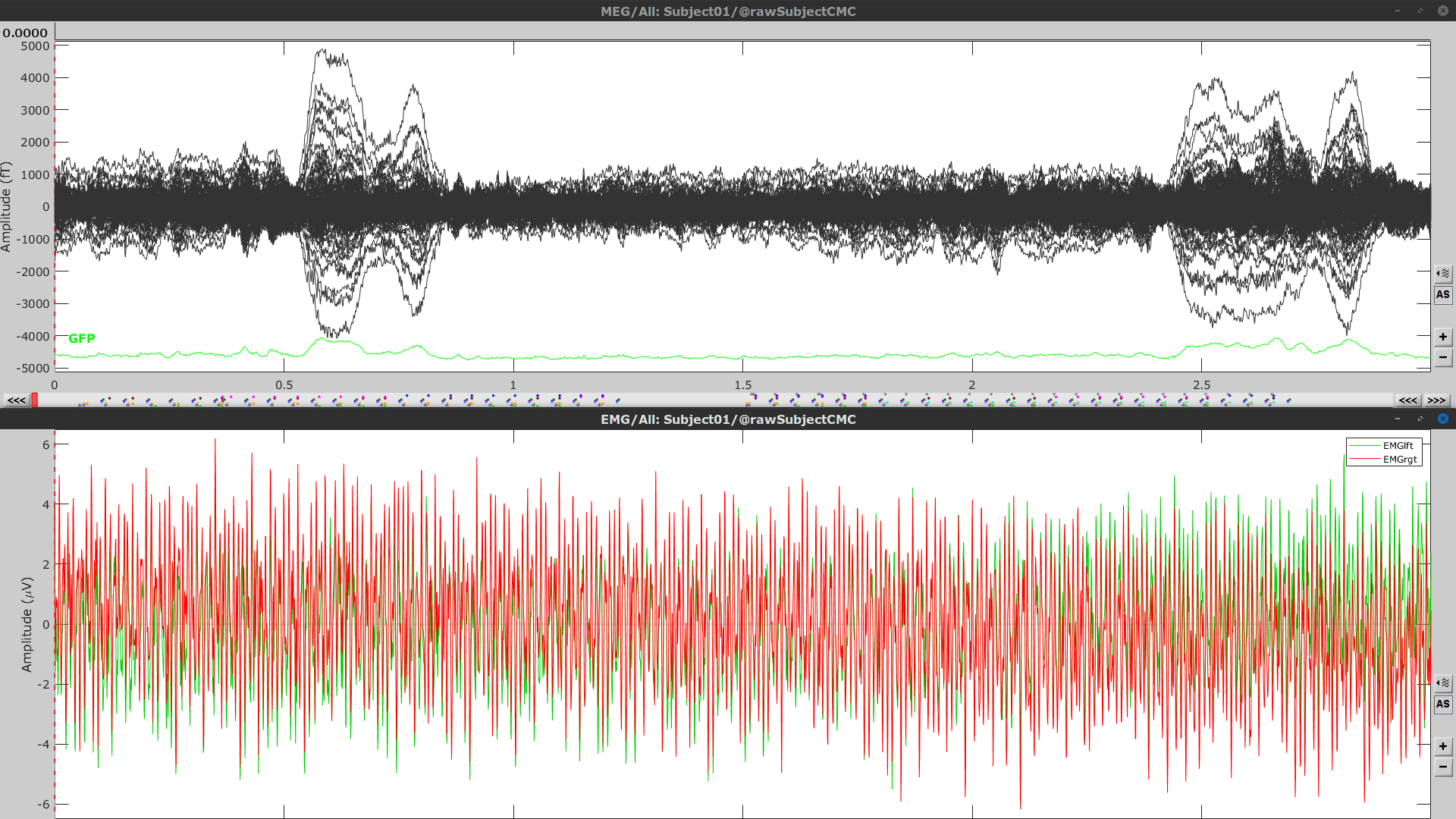
Right-click again on the Link to raw file node, then MEG > Display time series (or double-click on the node). This will open a new visualization window to explore data time series, also enabling the Time panel and the Record tab in the main Brainstorm window (see how to best use all controls in this panel and tab to explore data time series).
We will also display EMG traces by right-clicking on the Link to raw file node, then EMG > Display time series.
Event markers
The colored dots above the data time series indicate event markers (or triggers) saved with this dataset. The trial onset information of the left-wrist and right-wrist trials is saved in an auxiliary channel of the raw data named Stim. To add these markers, these events need to be decoded as follows:
While the time series figure is open, go to the Record tab and File > Read events from channel. From the options of the Read from channel process window, set Event channels = Stim, select Value, and click Run.
This procedure creates new event markers now shown in the Events section of the tab. along with previous event categories. In this tutorial, we will only use events U1 through U25, which correspond to how each of the 25 left-wrist trials had been encoded in the study. We will now delete other events of no interest, and merge the left trial events under a single event category, for convenience.
Delete other events: select the events to delete in the event box/list with Ctrl+click, then in the menu Events > Delete group and confirm. Alternatively, you can selected all events with Ctrl+A and deselect the U1 to U25 events by clicking on them.
To make sure we reproduce FieldTrip's tutorial, we need to reject trial #7: Select events U1 to U6 and U8 to U25, then from the Events menu, select Merge group and type ina new label (Left) to describe this is the left-wrist condition.
These events correspond to the beginning of 10-s trials of left-wrist movements. We will compute coherence over 1-s epochs over the first 8 s of each trial. To that purpose, we will now create extra events to define these epochs.
Duplicate 7 times the Left events by selecting Duplicate group in the Events menu. The groups Left_02 to Left_08 will be created.
For each copy of the Left events, we will add a time offset of 1 s for Left02, 2 s for Left03, and so on. Select the Left event group to add a 1,000 ms time offset, by going to the menu Events > Add time offset, enter 1,000 in the text box. Repeat for each other group, entering 2,000, then 3,000 etc.
Once done for Left_08, merge all these Left* events into a single Left category, and select Save modifications in the File menu in the Record tab.
Pre-process
In this tutorial will be analyzing only the Left trials. As such, in the following sections we will process only the first 330 s of the recordings.
The CTF MEG recordings in this dataset were not saved with the desired 3rd order compensation. To continue with the pre-processing we need to apply the compensation.
In the Process1 box: Drag and drop the Link to raw file node.
Run process Artifacts > Apply SSP & CTF compensation:
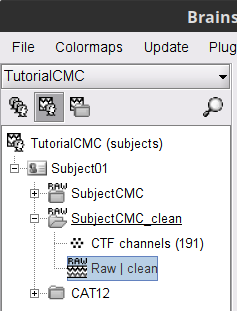
This process create the SubjectCMC_clean folder that contains a copy of the channel file and the raw recordings file Raw | clean, which is de exact copy of the original data but with the CTF compensation applied.
Power line artifacts
Let's start with locating the spectral components and impact of the power line noise in the MEG and EMG signals.
In the Process1 box: Drag and drop the Raw | clean node.
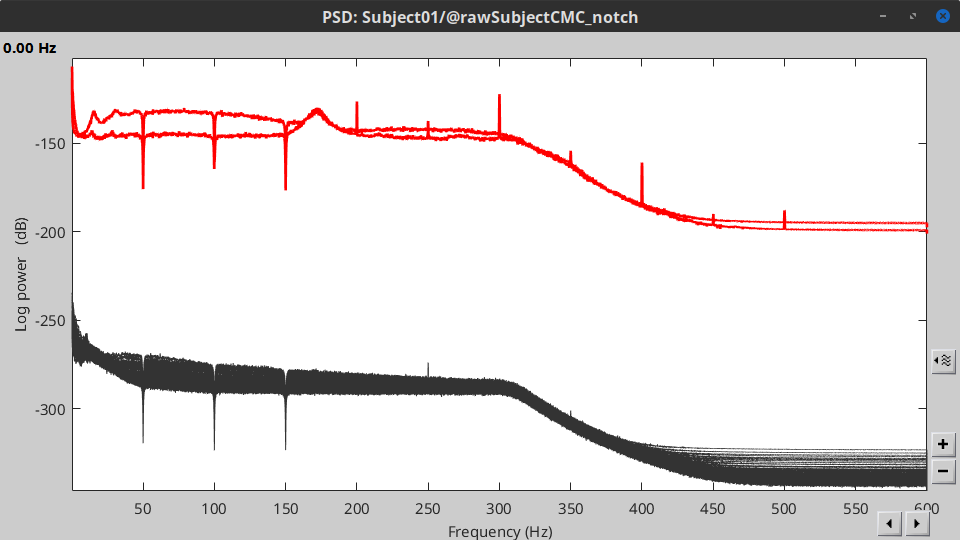
Run process Frequency > Power spectrum density (Welch):
Time window: 0 - 330 s
Window length=10 s
Overlap=50%
Sensor types=`MEG, EMG
Double-click on the new PSD file to display it.
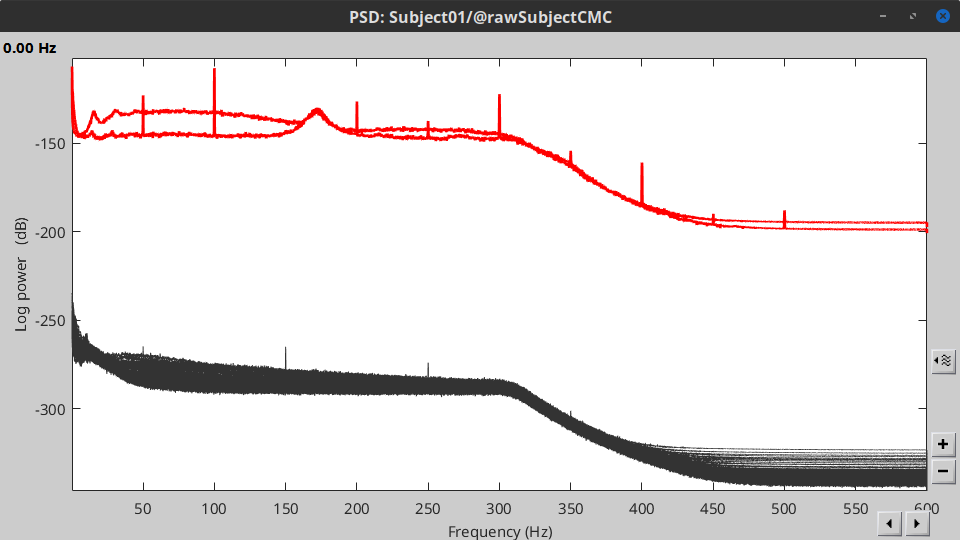
- The PSD shows two groups of sensors, EMG on top and MEG in the bottom. Also, there are peaks at 50Hz and its harmonics. We will use notch filters to remove the power line component and its first two components from the signals.
Run the process Pre-processing > Notch filter with:
Sensor types = MEG, EMG
Frequencies to remove (Hz) = 50, 100, 150
A new raw folder named SubjectCMC_clean_notch will appear in the database explorer. Compute the PSD for the filtered signals to verify effect of the notch filters. Remember to compute for the Time window from 0 to 330 s.
Pre-process EMG
Two of the typical pre-processing steps for EMG consist in high-pass filtering and rectifying.
In the Process1 box: drag and drop the Raw | notch(50Hz 100Hz 150Hz) recordings node.
Add the process Pre-process > Band-pass filter
Sensor types = EMG
Lower cutoff frequency = 10 Hz
Upper cutoff frequency = 0 Hz
Add the process Pre-process > Absolute values
Sensor types = EMG
- Run the pipeline
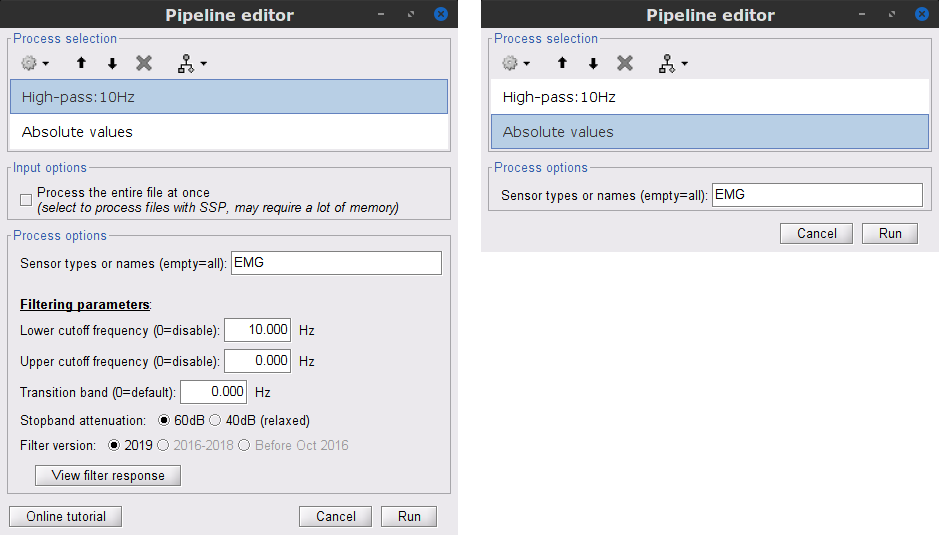
Once the pipeline ends, the new folders SubjectCMC_clean_notch_high and SubjectCMC_clean_notch_high_abs are added to the database explorer. To avoid any confusion later, we can delete folders that will not be needed.
Delete the conditions SubjectCMC_clean_notch and SubjectCMC_clean_notch_high. Select both folders containing and press Delete (or right-click File > Delete).
Pre-process MEG
After applying the notch filter to the MEG signals, we still need to remove other type of artifacts, we will perform:
Detection and removal of artifacts with SSP
Detection of segments with other artifacts
Detection and removal of artifacts with SSP
In the case of stereotypical artifacts, as it is the case of the eye blinks and heartbeats, it is possible to identify their characteristic spatial distribution, and then remove it from MEG signals with methods such as Signal-Space Projection (SSP). For more details, consult the tutorials on detection and removal of artifacts with SSP. The dataset of this tutorial contains an EOG channel but not ECG signal, thus will perform only removal of eye blinks.
Display the MEG and EOG time series. Right-click on the pre-processed (for EMG) continuous file Raw | clean | notch(... (in the SubjectCMC_clean_notch_high_abs folder) then MEG > Display time series and EOG > Display time series.
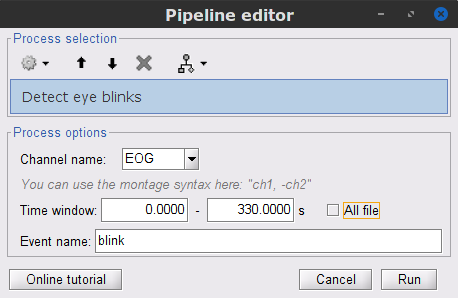
In the Events section of the Record tab, select Artifacts > Detect eye blinks, and use the parameters:
Channel name= EOG
Time window = 0 - 330 s
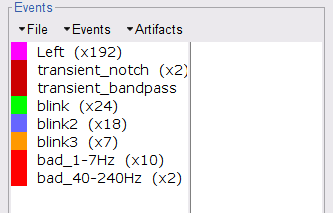
Event name = blink
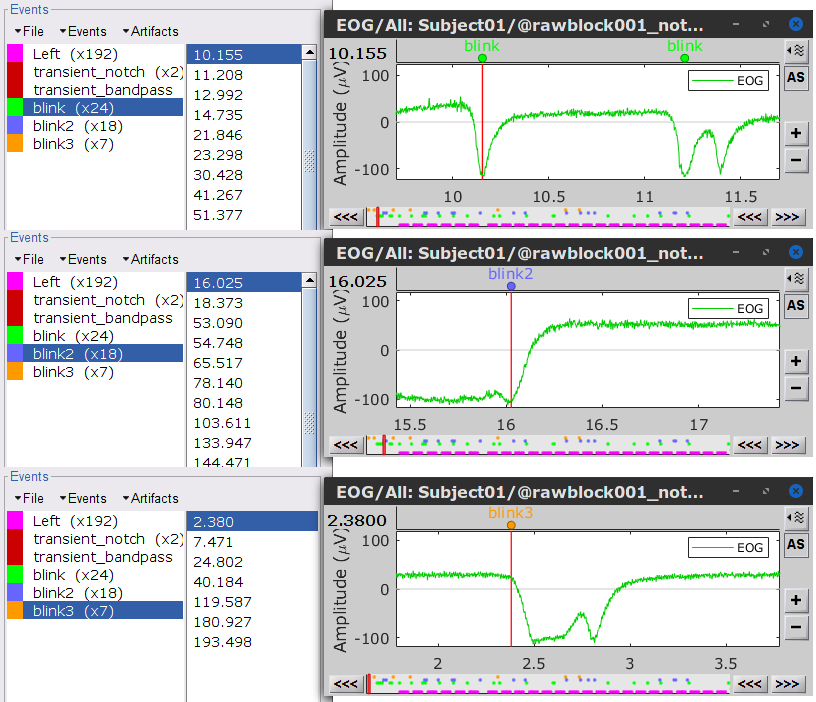
As result, there will be 3 blink event groups. Review the traces of EOG channels and the blink events to be sure the detected events make sense. Note that the blink group contains the real blinks, and blink2 and blink3 contain mostly saccades.
To remove blink artifacts with SSP go to Artifacts > SSP: Eye blinks, and use the parameters:
Event name=blink
Sensors=MEG
Check Compute using existing SSP/ICA projectors
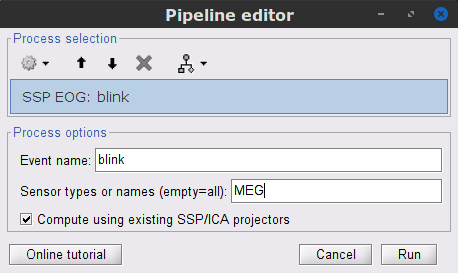
- Display the time series and topographies for the first two components. Only the first one is clearly related to blink artifacts. Select only component #1 for removal.
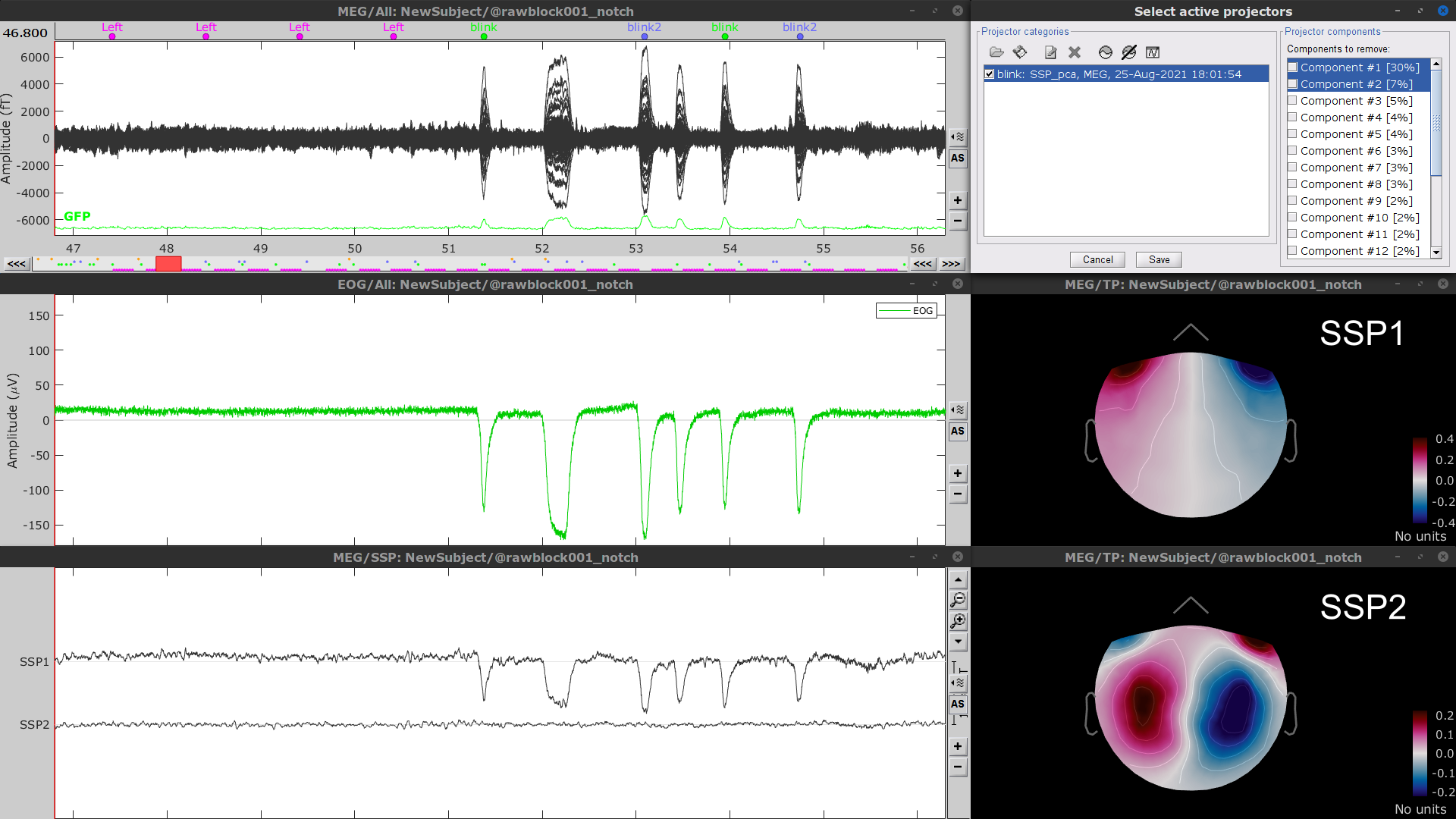
Follow the same procedure for the other blink events (blink2 and blink3). Note that none of first two components for the remaining blink events is clearly related to a ocular artifacts. This figure shows the first two components for the blink2 group.
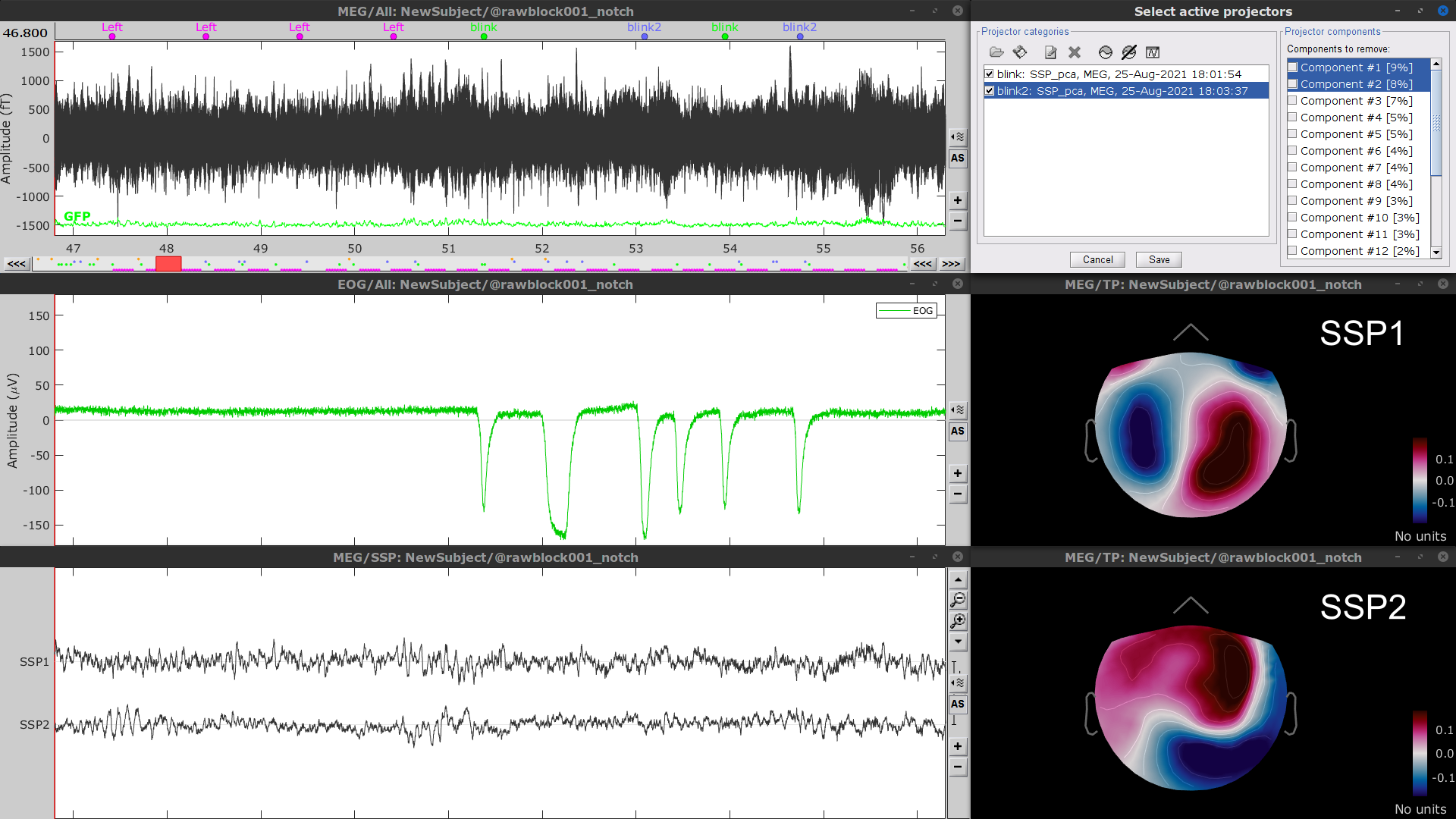
In this case, it is safer to unselect the blink2 and blink3 groups, rather than removing spatial components that we are not sure to identify.
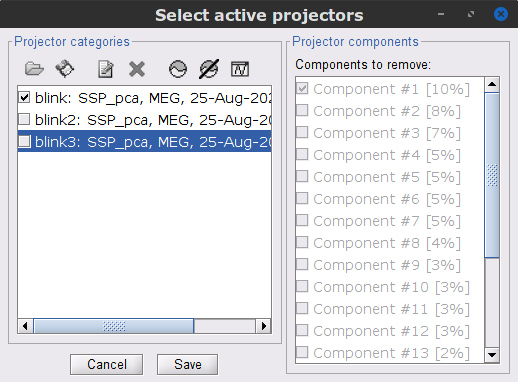
- Close all the figures
Detection of segments with other artifacts
Here we will used automatic detection of artifacts. It aims to identify typical artifacts such as the ones related to eye movements, subject movement and muscle contractions.
Display the MEG and EOG time series. In the Record tab, select Artifacts > Detect other artifacts, use the following parameters:
Time window = 0 - 330 s
Sensor types=MEG
Sensitivity=3
Check both frequency bands 1-7 Hz and 40-240 Hz
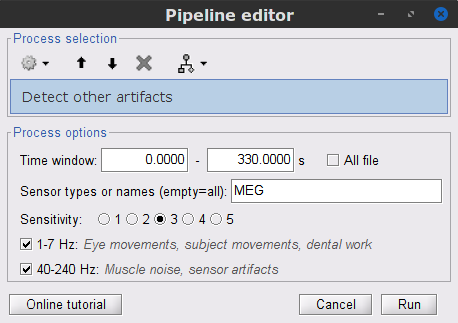
While this process can help identify segments with artifacts in the signals, it is still advised to review the selected segments. After a quick browse, it can be noticed that the selected segments indeed correspond to irregularities in the MEG signal. Then, we will label these events are bad.
Select the 1-7Hz and 40-240Hz event groups and use the menu Events > Mark group as bad. Alternatively, you can rename the events and add the tag bad_ in their name, it would have the same effect.
- Close all the figures, and save the modifications.
Importing the recordings
At this point we have finished with the pre-processing of our EMG and MEG recordings. Many operations operations can only be applied to short segments of recordings that have been imported in the database. We refer to these as epochs or trials. Thus, the next step is to import the data taking into account the Left events.
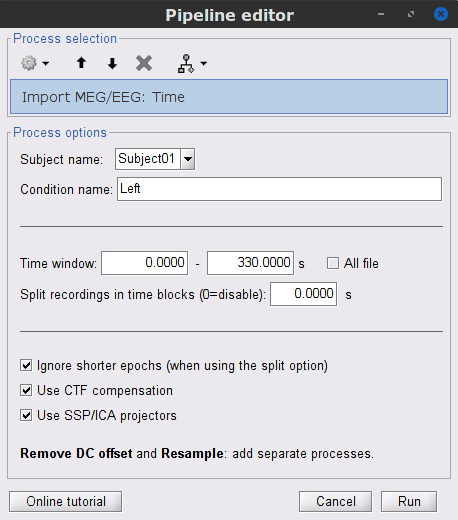
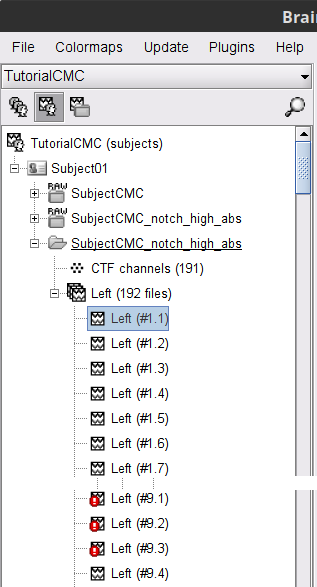
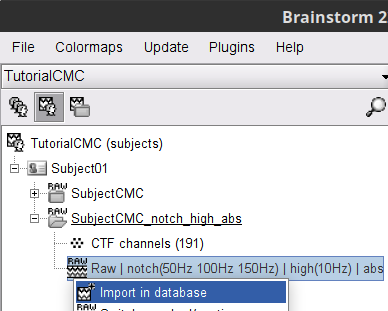
Right-click on the filtered continuous file Raw | clean | notch(... (in the SubjectCMC_clean_notch_high_abs condition), then Import in database.
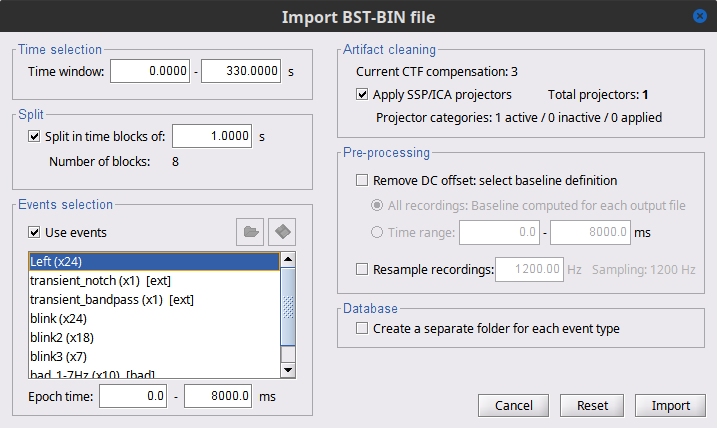
- Set the following parameters:
Time window = 0 - 330 s
Check Use events and highlight the Left(x192) event group
Epoch time = 0 - 1000 ms
Check Apply SSP/ICA projectors
Check Remove DC offset and select All recordings
The new folder SubjectCMC_clean_notch_high_abs appears for Subject01. It contains a copy of the channel file in the continuous file, and the Left trial group. By expanding the trial group, we can notice that there are trials marked with an interrogation sign in a red circle (ICON). These bad trials are the ones that were overlapped with the bad segments identified in the previous section. All the bad trials are automatically ignored in the Process1 and Process2 tabs.
Coherence (sensor level)
Once we have imported the trials, we will compute the magnitude square coherence (MSC) between the left EMG signal and the signals from each of the MEG sensors.
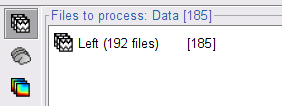
In the Process1 box, drag and drop the Left (192 files) trial group. Note that the number between square brackets is [185], as the 7 bad trials are ignored.
To compute the coherence between EMG and MEG signals. Run the process Connectivity > Coherence 1xN [2021] with the following parameters:
Time window = 0 - 1000 ms or check All file
Source channel = EMGlft
Do not check Include bad channels nor Remove evoke response
Magnitude squared coherence
Window length for PSD estimation = 0.5 s
Overlap for PSD estimation = 50%
Highest frequency of interest = 80 Hz
Average cross-spectra of input files (one output file)
More details on the Coherence process can be found in the ?connectivity tutorial.
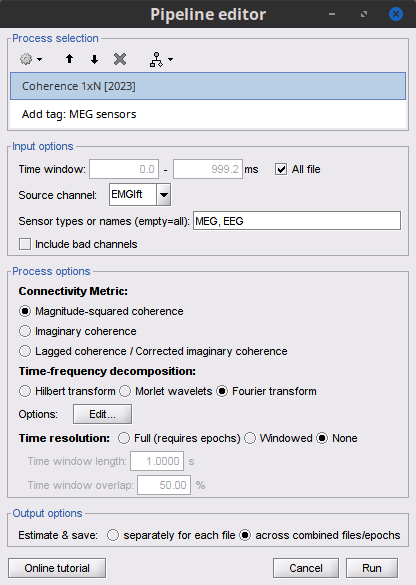
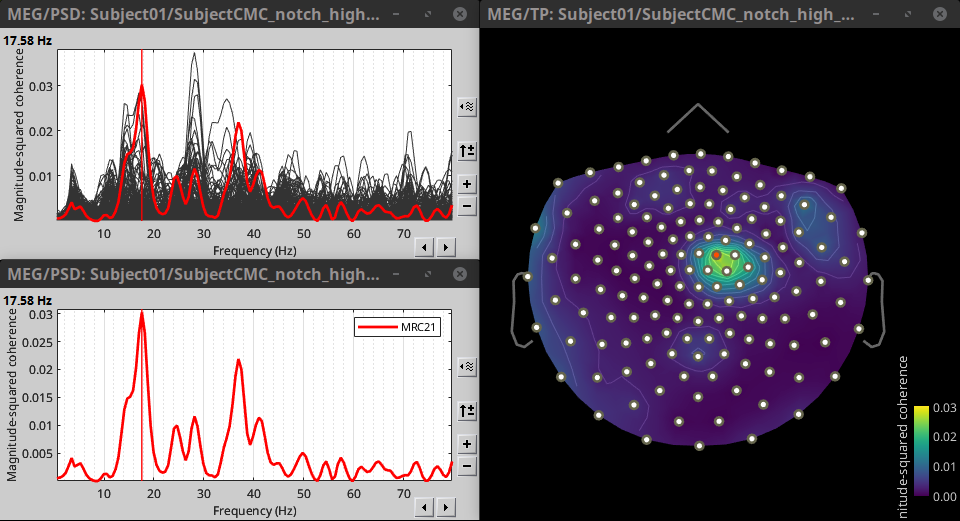
Double-click on the resulting node mscohere(0.6Hz,555win): EMGlft to display the MSC spectra. Click on the maximum peak in the 15 to 20 Hz range, and press Enter to plot it in a new figure. This spectrum corresponds to channel MRC21, and has its peak at 17.58 Hz. You can also use the frequency slider (below the Time panel) to explore the spectral representations.
* Right-click on the spectrum and select 2D Sensor cap for a spatial visualization of the coherence results, alternatively, the short cut Ctrl-T can be used. Once the 2D Sensor cap is show, the sensor locations can be displayed with right-click then Channels > Display sensors or the shortcut Ctrl-E.
The results above are based in the identification of single peak, as alternative we can average the MSC in a given frequency band (15 - 20 Hz), and observe its topographical distribution.
In the Process1 box, drag-and-drop the mscohere(0.6Hz,555win): EMGlft node, and add the process Frequency > Group in time or frequency bands with the parameters:
Select Group by frequency
Type cmc_band / 15, 20 / mean in the text box.
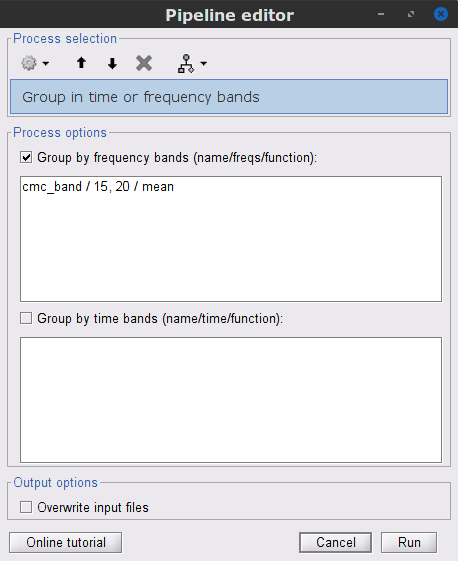
The resulting file mscohere(0.6Hz,555win): EMGlft | tfbands has only one MSC value for each sensor (the average in the 15-20 Hz band). Thus, it is more useful to display the result in a spatial representation. Brainstorm provides 3 spatial representations: 2D Sensor cap, 2D Sensor cap and 2D Disk, which are accessible with right-click on the MSC node. Sensor MRC21 is selected as reference.
In agreement with the literature, we observe higher MSC values between the EMG signal and the MEG signal for MEG sensors over the contralateral primary motor cortex in the beta band range. In the next sections we will perform source estimation and compute coherence in the source level.
Source analysis
In this tutorial we will perform source modelling using the distributed model approach for two sources spaces: cortex surface and MRI volume. In the first one the location of the sources is constrained to the cortical surface obtained when the subject anatomy was imported. For the second source space, the sources are uniformly distributed in the entire brain volume. Before estimating the brain sources, we need to compute head model and the noise covariance. Note that a head model is required for each source space.
Head model
The head model describes how neural electric currents produce magnetic fields and differences in electrical potentials at external sensors, given the different head tissues. This model is independent of sensor recordings. See the head model tutorial for more details. Each source space, requires its own head model.
Cortex surface
In the SubjectCMC_clean_notch_high_abs, right-click the CTF channels (191) node and select Compute head model. Keep the default options:
Comment = Overlapping spheres (surface)
Source space = Cortex surface
Forward model = Overlapping spheres.
Keep in mind that the number of sources (vertices) in this head model is 10,000, and was defined when when the subject anatomy was imported.
The (ICON) Overlapping spheres (surface) head model will appear in the database explorer.
MRI volume
In the SubjectCMC_clean_notch_high_abs, right-click the CTF channels (191) node and select Compute head model. Keep the default options:
Comment = Overlapping spheres (volume)
Source space = MRI volume
Forward model = Overlapping spheres.
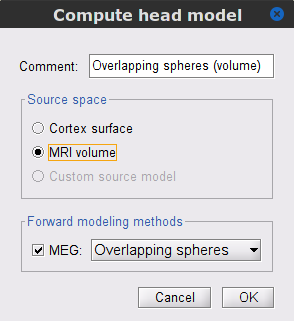
The Volume source grid window pop-up, to define the volume grid. Use the following parameters, that will lead to an estimated number of 12,200 grid points.
Select Regular grid and Brain
Grid resolution = 5 mm
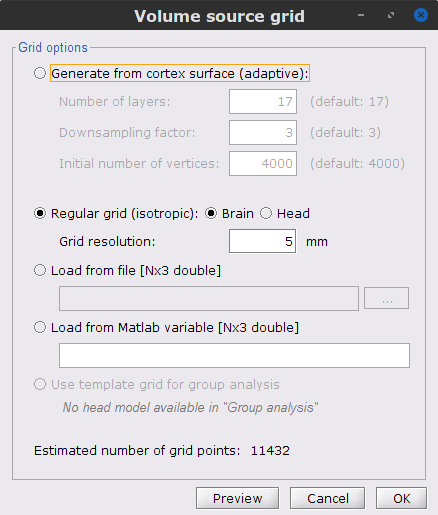
The Overlapping spheres (volume) node will be added to the database explorer. The green color indicates the default head model for the folder.
Noise covariance
For MEG recordings it is recommended to derive the noise covariance from empty room recordings. However, as we do not have those recordings in the dataset, we can compute the noise covariance from the MEG signals before the trials. See the noise covariance tutorial for more details.
In the raw SubjectCMC_clean_notch_high_abs, right-click the Raw | clean | notch(... node and select Noise covariance > Compute from recordings. As parameters select:
Baseline from 18 - 30 s
Select the Block by block option.
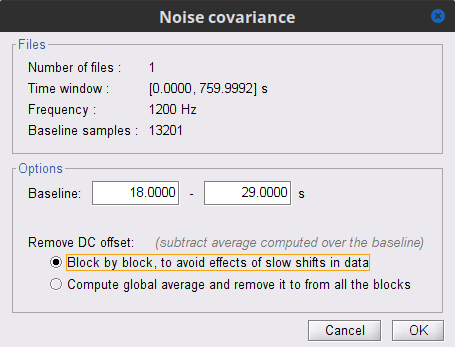
Lastly, copy the Noise covariance node to the SubjectCMC_clean_notch_high_abs folder with the head model. This can be done with the shortcuts Ctrl-C and Ctrl-V.
Source estimation
Noe that the head model(s) and noise covariance have been computed, we can use the minimum norm imaging method to solve the inverse problem. The result is a linear inversion kernel, that estimates the source brain activity that gives origin to the observed recordings in the sensors. Note that, an inversion kernel is obtained for each of the head models: surface and volume. See the source estimation tutorial for more details.
Cortex surface
Compute the inversion kernel, right-click in the Overlapping spheres (surface) head model and select Compute sources [2018]. With the parameters:
Minimum norm imaging
Current density map
Unconstrained
Comment = MN: MEG (surface)
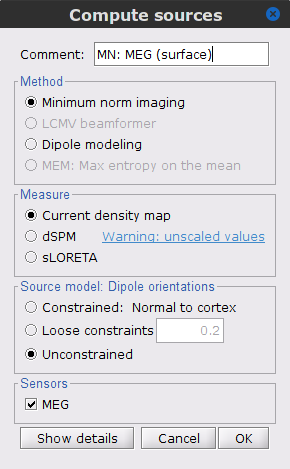
The inversion kernel (ICON) MN: MEG (surface)(Unconstr) 2018 is created, and added to the database explorer.
MRI volume
Compute the inversion kernel, right-click in the Overlapping spheres (volume) head model and select Compute sources [2018]. With the parameters:
Minimum norm imaging
Current density map
Unconstrained
Comment = MN: MEG (volume)
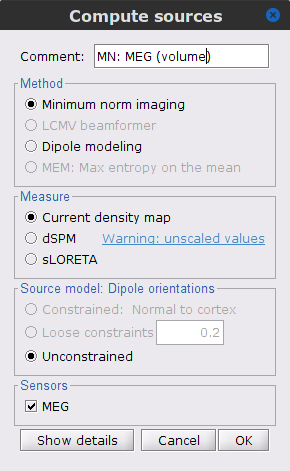
The inversion kernel (ICON) MN: MEG (volume)(Unconstr) 2018 is created, and added to the database explorer. The green color in the name indicates the current default head model. In addition, note that each trial has now two associated source link (ICON) nodes. One obtained with the MN: MEG (surface)(Unconstr) 2018 kernel and the other obtained with the MN: MEG (volume)(Unconstr) 2018 kernel.
Scouts
From the head model section, we notice that the cortex and volume grid have around 10,000 vertices each, thus as many sources were estimated. As such, it is not practical to compute coherence between the left EMG signal and the signal of each source. A way to address this issue is with the use of regions of interest also known as scouts. Thus, there are surface scouts and volume scouts. Let's define scouts for the different source spaces.
Surface scouts
In the source link MN: MEG (surface)(Unconstr) 2018 node for one of the trials, right-click and select Cortical activations > Display on cortex. In the Surface tab, set the Amplitude slider to 100% to hide all the sources.
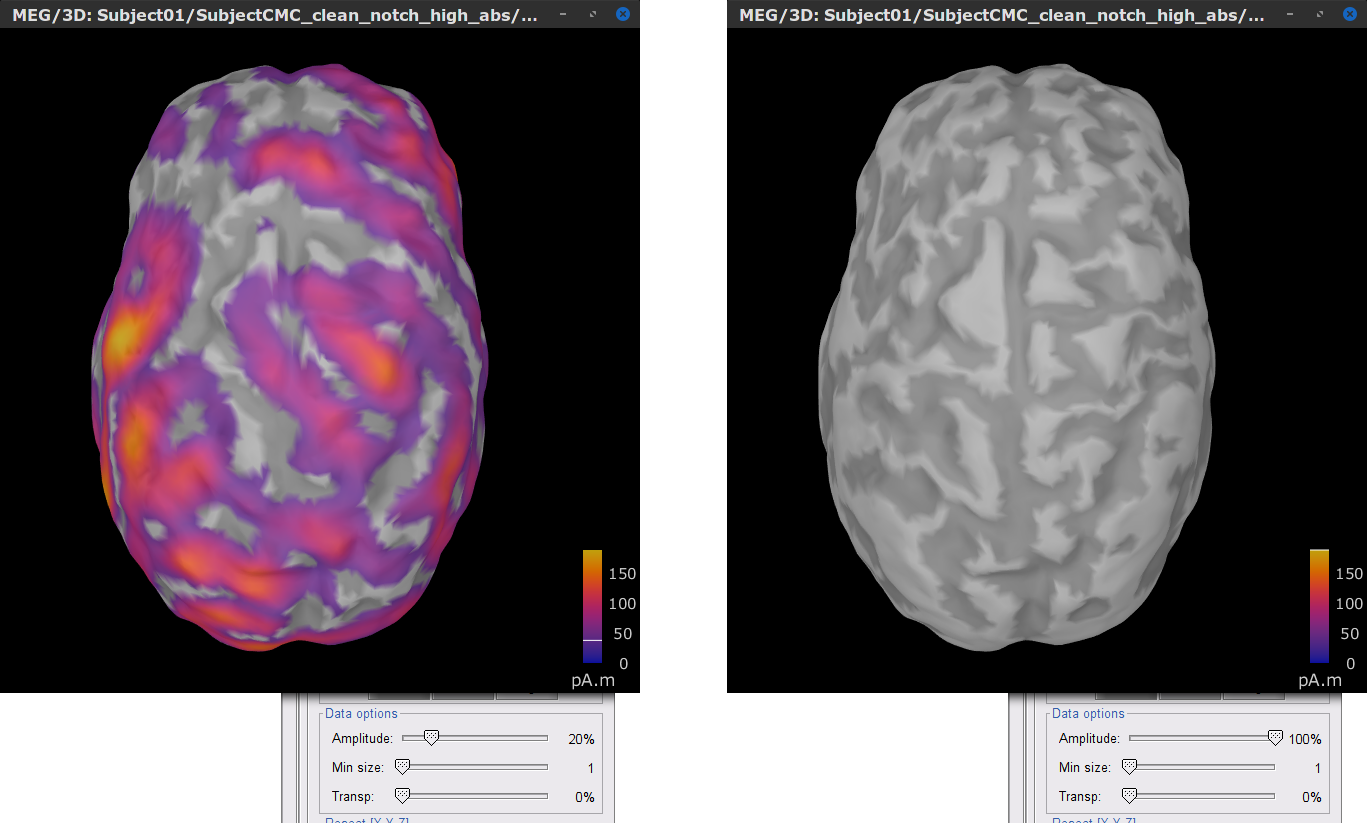
In the Scout tab, select the menu Atlas > From subject anatomy > AAL3 (MNI-linear). This will create the From volume: AAL3 set of surface scouts. By clicking in the different scouts, at the bottom of the list, the number of vertices it contains and the approximate area in cm2 is shown. Activate the (ICON) Show only the selected scouts option to narrow down the shown scouts.
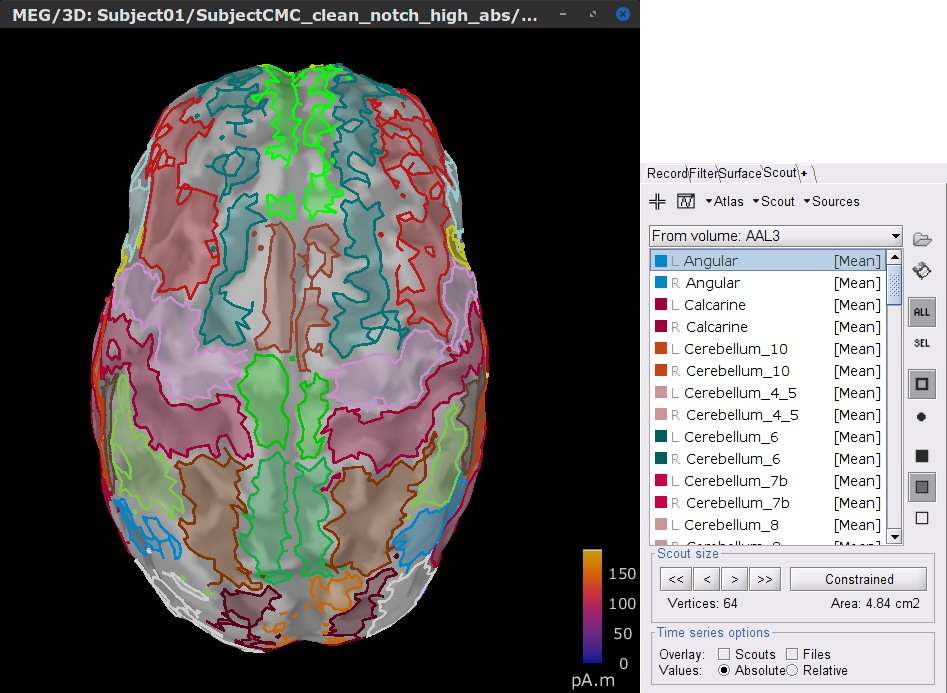
[TODO] A note, that the definition of scouts is far from perfect, but can give us a good idea of the surface projections of the MNI parcellations (described in the importing anatomy section).
- Close the figure.
Volume scouts
In the source link MN: MEG (volume)(Unconstr) 2018 node for one of the trials, right-click and select Cortical activations > Display on MRI (3D): Subject CMC. In the Surface tab, set the Amplitude slider to 100% to hide all the sources.
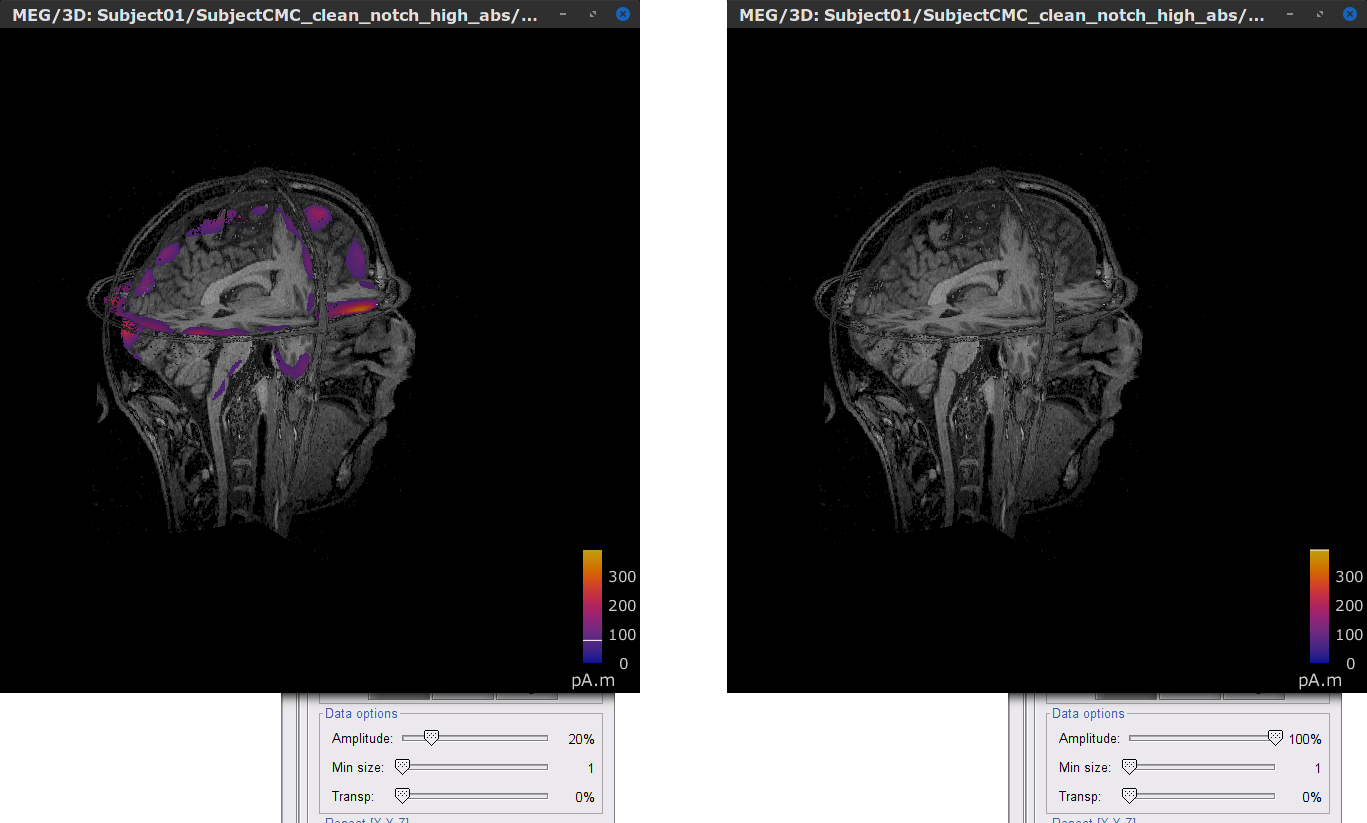
In the Scout tab, select the menu Atlas > From subject anatomy > AAL3 (MNI-linear). This will create the Volume 12203: AAL3 set of volume scouts. By clicking in the different scouts, at the bottom of the list, the number of vertices it contains and the approximate volume in cm3 is shown. Activate the (ICON) Show only the selected scouts option to narrow down the shown scouts.
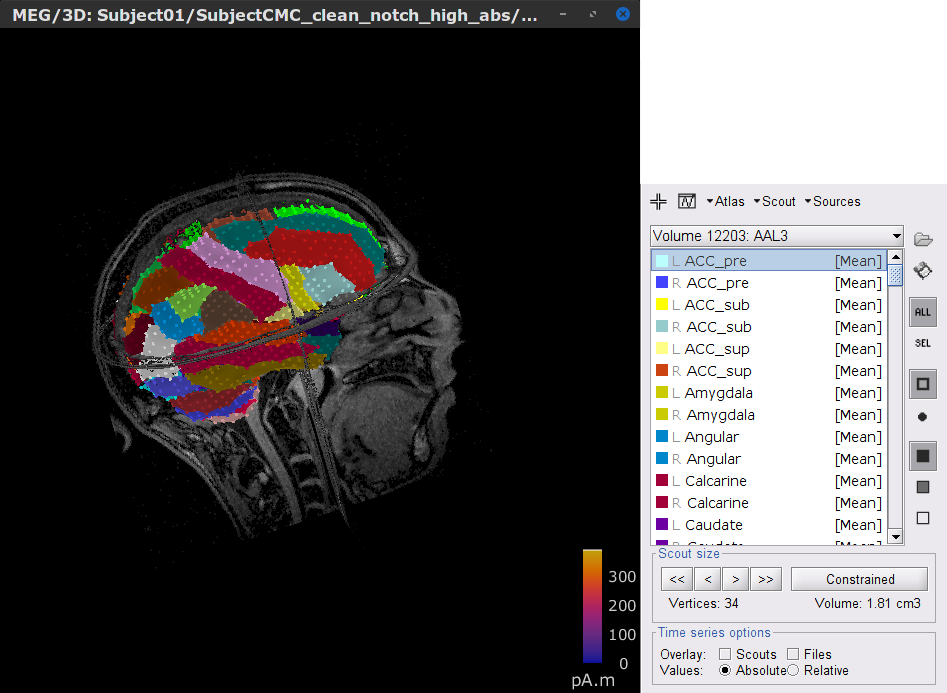
- Close the figure.
Coherence (source level)
Coherence in the source level is computed between a sensor signal (EMG) and source signals in the (surface or volume) scouts.
Coherence with surface scouts
To facilitate the selection of the indicated files to compute this coherence, let's search in the database the recordings and the source link files obtained wit the MN: MEG (surface)(Unconstr) 2018 kernel.
Click on the magnifying glass (ICON) above the database explorer to open up the search dialog, and select New search.
Set the search query to look for files that are named (surface) or are named Left. This is done with the following configuration.
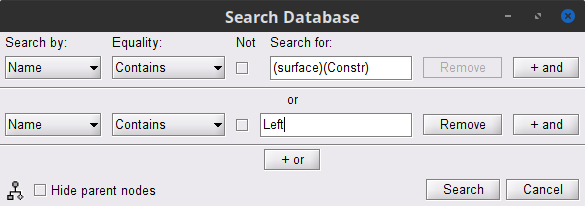
By performing the search, a new tab called (surface) appears in the database explorer. This new tab contains the recordings and ONLY the source link for the surface space.
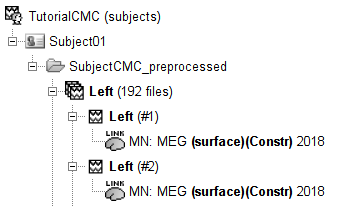
Change to the Process2 tab, and drag-and-drop the Left (192 files) trial group into the Files A and into the Files B boxes. And select Process recordings for Files A, and Process sources for Files B. Note that blue labels over the Files A and the Files B boxes indicate that there are 185 files per box.
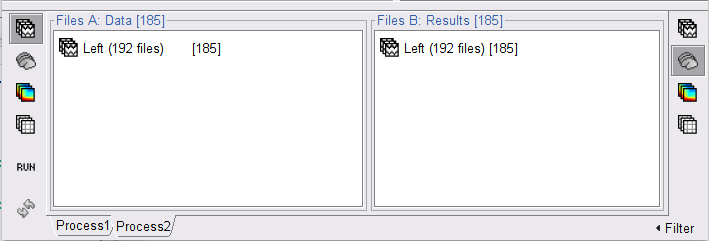
Open the Pipeline editor and add two process:
Add the process Connectivity > Coherence AxB [2021] with the following parameters:
Time window = 0 - 1000 ms or check All file
Source channel (A) = EMGlft
Check Use scouts (B)
Select From volume: AAL3 in the drop-down list (these are surface scouts)
Select all the scouts (shortcut Ctrl-A)
Scout function = Mean
When to apply = Before
Do not Remove evoked responses from each trial
Magnitude squared coherence, Window length = 0.5 s
Overlap = 50%
Highest frequency = 80 Hz
Average cross-spectra.
Add the process File > Add tag with the following parameters:
Tag to add = (surface)
Select Add to file name
- Run the pipeline
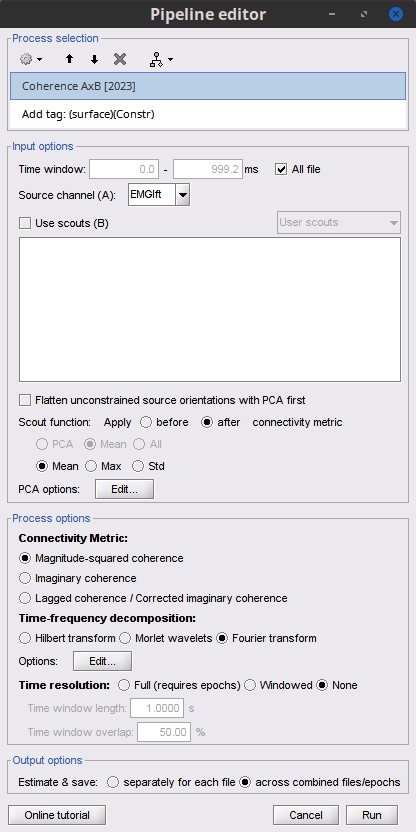
Double-click on the resulting node mscohere(0.6Hz,555win): Left (#1) | (surface) to display the coherence spectra. Also open the result node as image with Display image in its context menu.
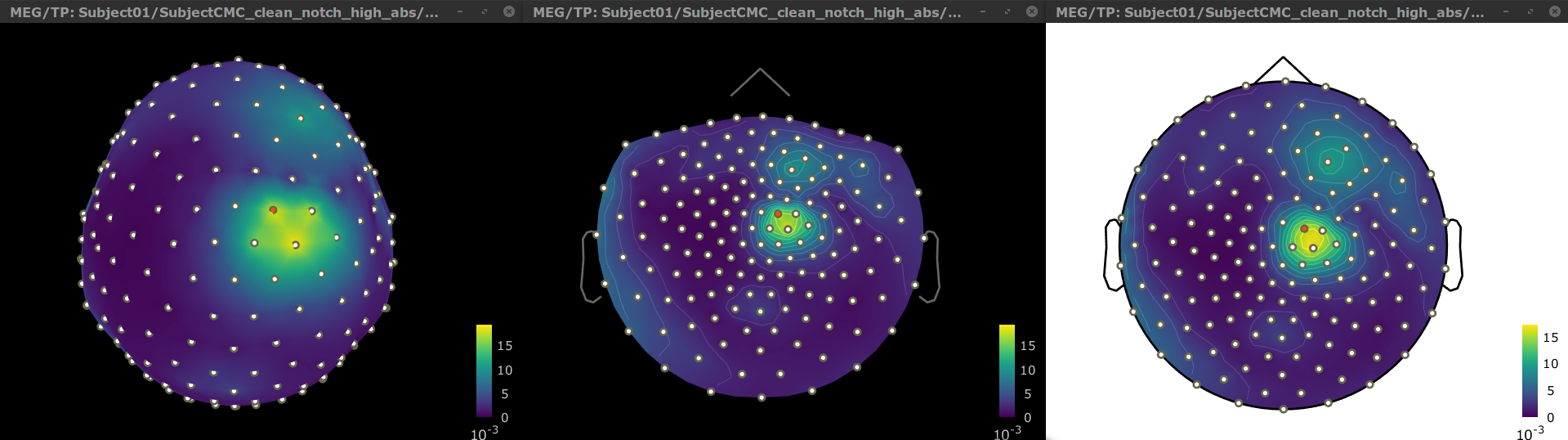
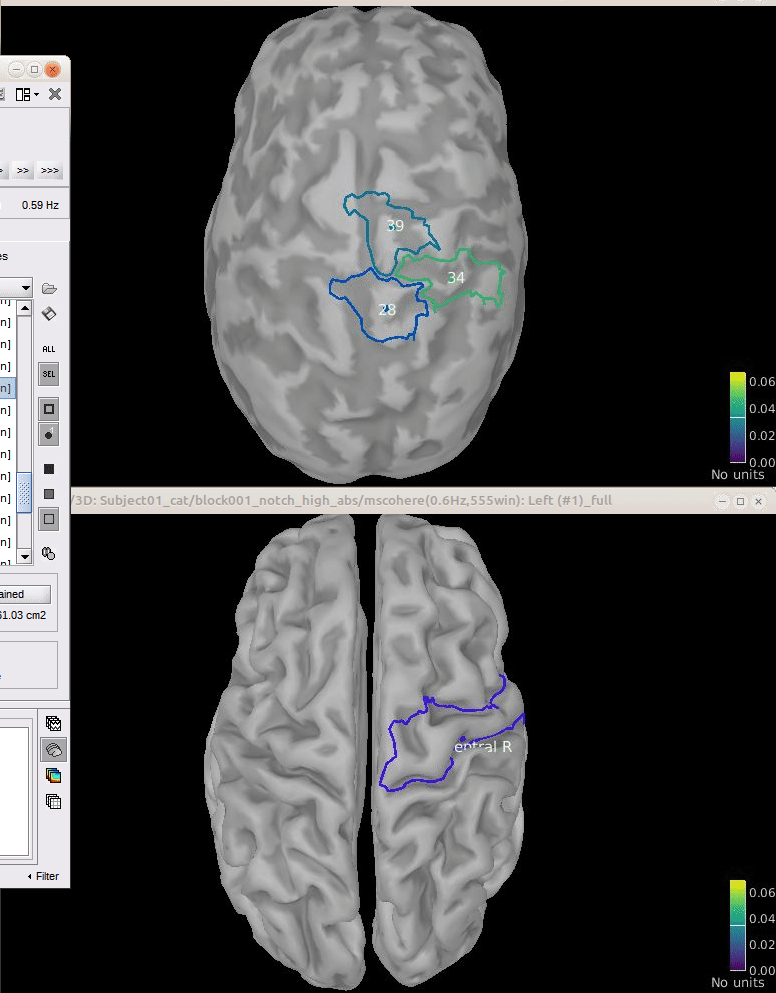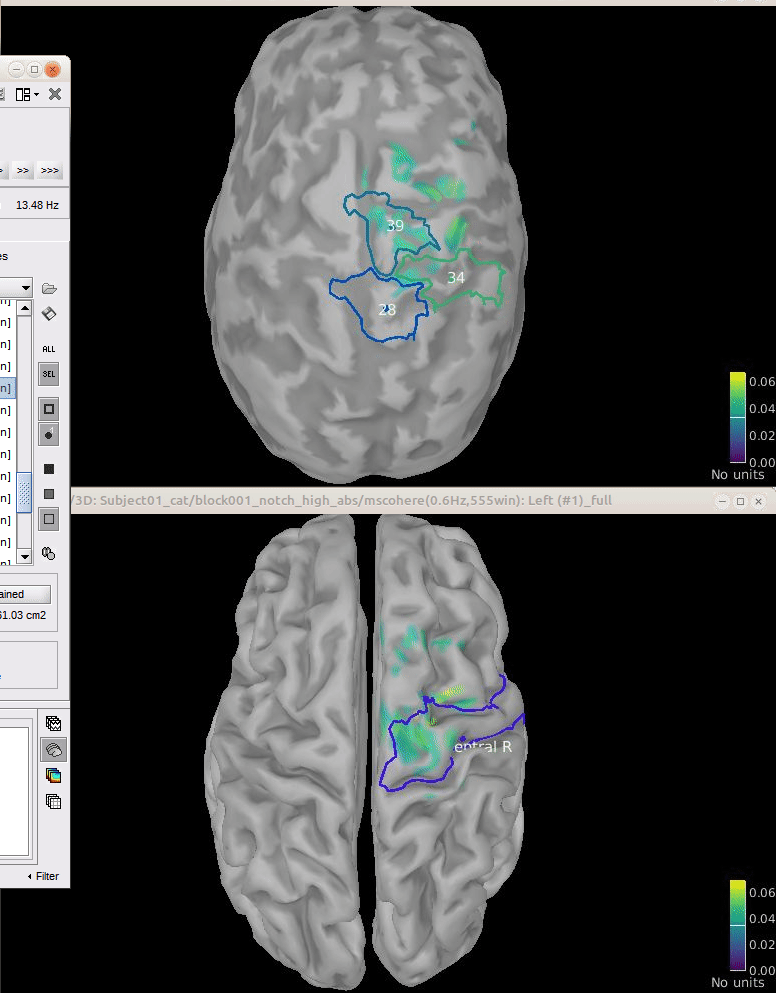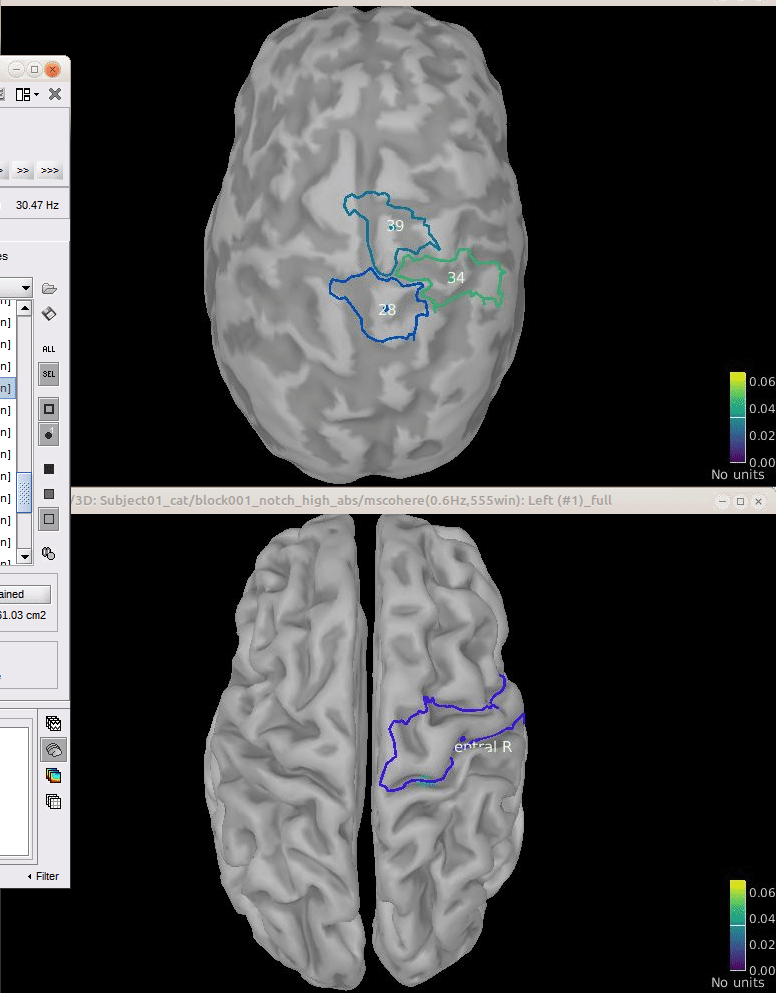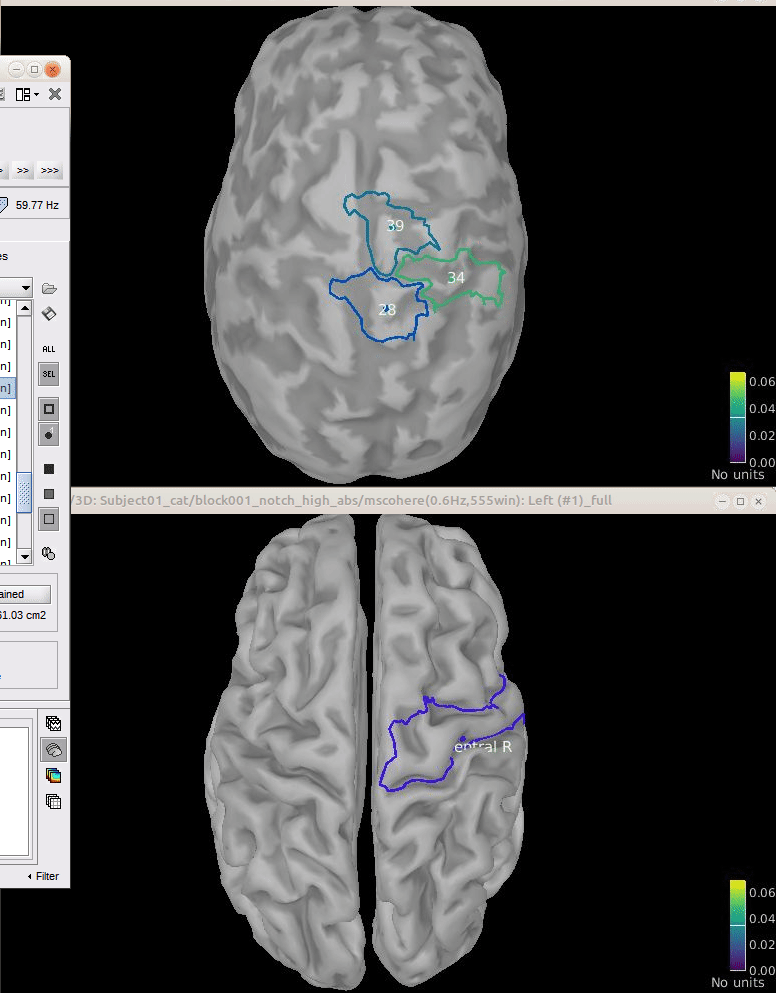
To verify the location of the scouts on the cortex surface, double-click one of the (surface) source link for any of the trials. In the Surface tab, set the Amplitude threshold to 100% to hide all the cortical activations. Lastly, in the Scouts tab, select the From volume: AAL3 atlas in the drop-down list, select Show only the selected scouts and the Show/hide the scout labels. Note that the plots are linked by the scout selected in the image representation of the coherence results.
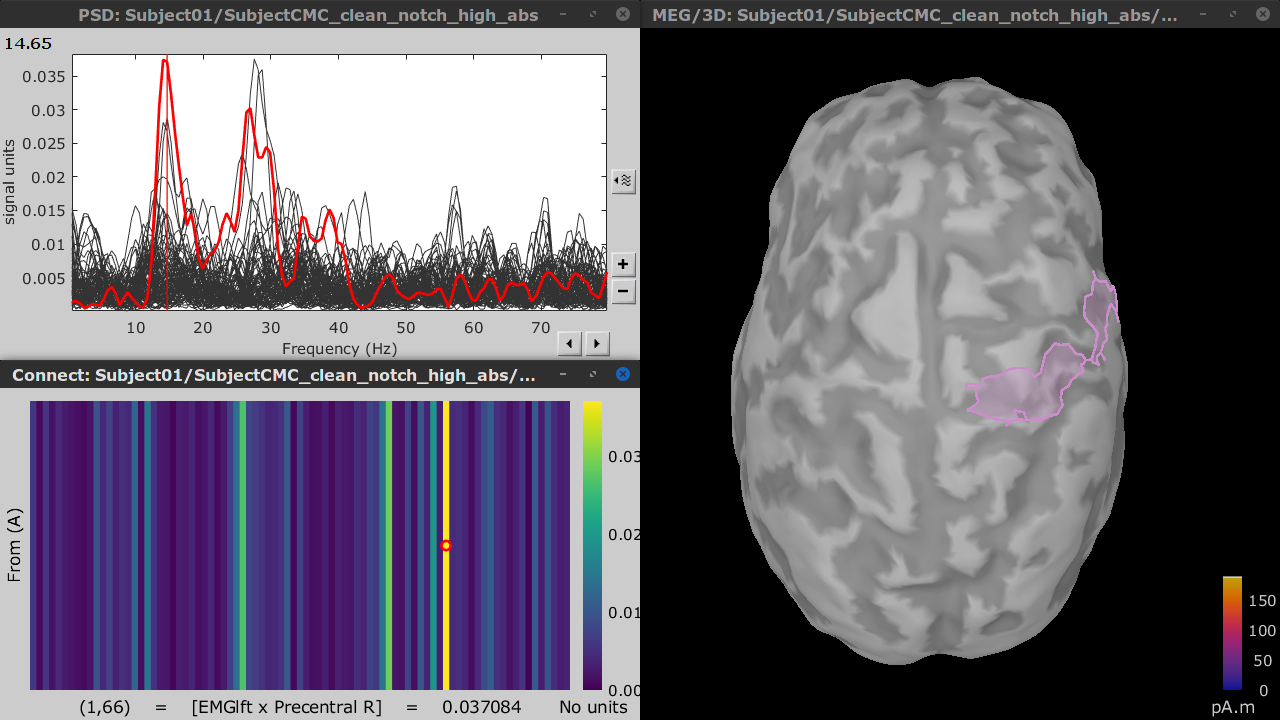
From the results we can see that the peak at 14.65 Hz corresponds to the Precentral R scout, which encompasses the right primary motor cortex, as expected. These results are inline with the ones in the literature.
Coherence with volume scouts
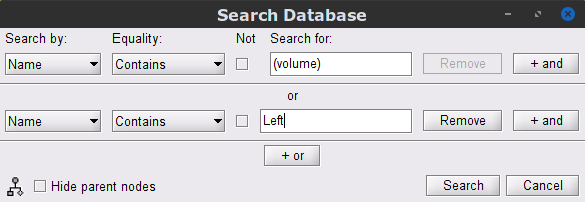
Similar to coherence with surface scouts, a search is needed to select the recordings and the source link files obtained wit the MN: MEG (volume)(Unconstr) 2018 kernel.
Click on the magnifying glass (ICON) above the database explorer to open up the search dialog, and select New search. Set the search query to look for files that are named (volume) or are named Left. This is done with the following configuration.
Change to the Process2 tab, and drag-and-drop the Left (192 files) trial group into the Files A and into the Files B boxes. And select Process recordings for Files A, and Process sources for Files B.

Open the Pipeline editor and add two process:
Add the process Connectivity > Coherence AxB [2021] with the following parameters:
Time window = 0 - 1000 ms or check All file
Source channel (A) = EMGlft
Check Use scouts (B)
Select Volume 12203: AAL3 in the drop-down list (these are volume scouts)
Select all the scouts (shortcut Ctrl-A)
Scout function = Mean
When to apply = Before
Do not Remove evoked responses from each trial
Magnitude squared coherence, Window length = 0.5 s
Overlap = 50%
Highest frequency = 80 Hz
Average cross-spectra.
Add the process File > Add tag with the following parameters:
Tag to add = (surface)
Select Add to file name
- Run the pipeline
Double-click on the resulting node mscohere(0.6Hz,555win): Left (#1) | (volume) to display the coherence spectra. Also open the result node as image with Display image in its context menu.
To verify the location of the scouts on the cortex surface, open one of the (volume) source link for any of the trials with the as MRI (3D), in the context menu select Display cortical activations > Display on MRI (3D): SubjectCMC . In the Surface tab, set the Amplitude threshold to 100% to hide all the cortical activations. Lastly, in the Scouts tab, select the Volume 12203: AAL3 atlas in the drop-down list, select the Show only the selected scouts and the Show/hide the scout labels. Note that the plots are linked by the scout selected in the image representation of the coherence results.
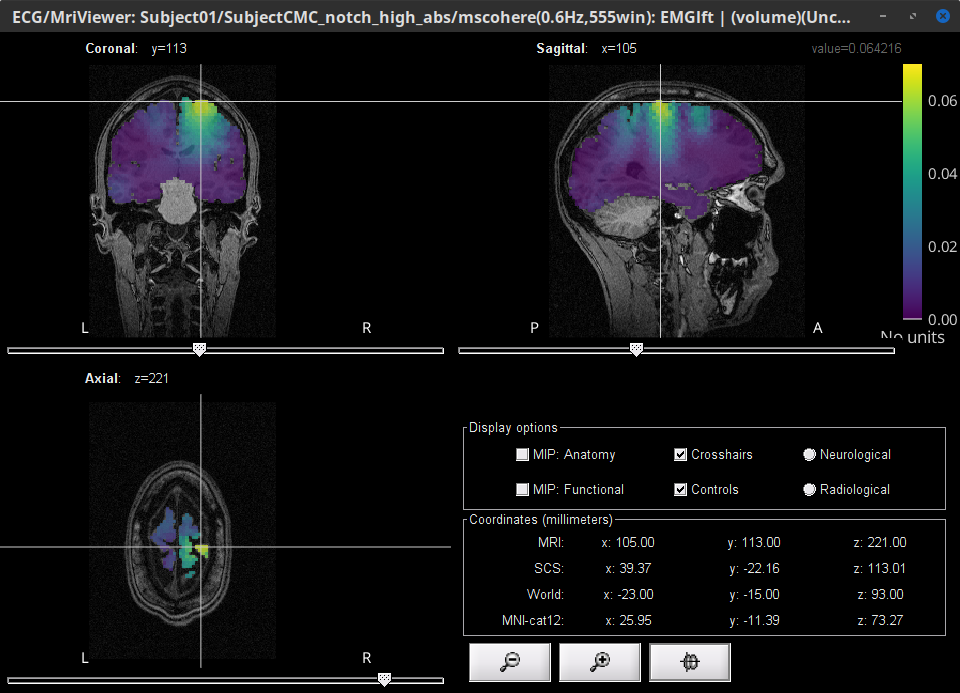
From the results we can see that the peak at 14.65 Hz corresponds to the Precentral R scout, which encompasses the right primary motor cortex, as expected. These results are inline with the ones in the literature.
Coherence with all sources (no scouts)
- We could downsample the surface and create a more sparse volume grid
- OR
- Refactor the coherence process to accumulate the auto- and cross-spectra outside of the function
- OR
Comparison of cortex surface with FieldTrip and CAT
[TO DISCUSS among authors] This image and GIF are just for reference. They were obtained with all the surface sources using ?FieldTrip and CAT derived surfaces.
Comparison for 14.65 Hz
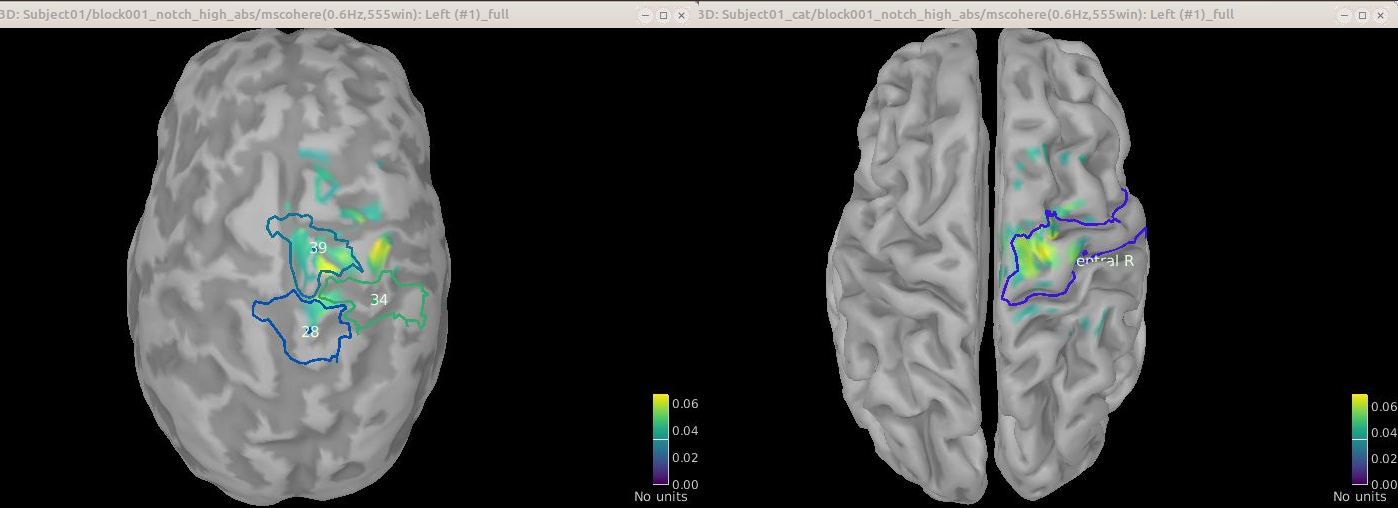
Sweeping from 0 to 80 Hz
Script
[TO DO] Once we agree on all the steps above.
Additional documentation
Articles
Conway BA, Halliday DM, Farmer SF, Shahani U, Maas P, Weir AI, et al.
Synchronization between motor cortex and spinal motoneuronal pool during the performance of a maintained motor task in man.
The Journal of Physiology. 1995 Dec 15;489(3):917–24.Kilner JM, Baker SN, Salenius S, Hari R, Lemon RN.
Human Cortical Muscle Coherence Is Directly Related to Specific Motor Parameters.
J Neurosci. 2000 Dec 1;20(23):8838–45.Liu J, Sheng Y, Liu H.
https://doi.org/10.3389/fnhum.2019.00100Corticomuscular%20Coherence%20and%20Its%20Applications:%20A%20Review. Front Hum Neurosci. 2019 Mar 20;13:100.
Tutorials
Tutorial: Volume source estimation
Tutorial: Functional connectivity
Forum discussions
[TO DO] Find relevant Forum posts.