|
Size: 47669
Comment:
|
Size: 34317
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| '''UNDER CONSTRUCTION''' |
|
| Line 18: | Line 16: |
| * '''SPM''': If you are running Brainstorm from the Matlab environment, for the coregistration between pre- and post-implantation MRI volumes, you need to have the [[http://www.fil.ion.ucl.ac.uk/spm/software/download/|SPM12 toolbox]] installed on your computer and added to your Matlab path. <<BR>>No need to worry about SPM if you are using the stand-alone compiled version of Brainstorm: all the needed SPM scripts have been compiled and included in this executable. * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and download the file: '''sample_ecog.zip''' * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder) * Start Brainstorm (Matlab scripts or stand-alone version). * Select the menu File > Create new protocol. Name it "'''TutorialEcog'''" and select the options: * "'''No, use individual anatomy'''", * "'''No, use one channel file per acquisition run'''". |
* '''SPM''': The coregistration between pre- and post-implantation MRI volumes requires the SPM12 toolbox. If you are running Brainstorm from a Matlab installation, SPM can be installed or configured as a [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|plugin]]: menu Plugins > SPM12 > Install. If you are using the stand-alone compiled version of Brainstorm: all the needed SPM scripts have been compiled and included in this executable. * '''Download the dataset''': * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and get the file: '''sample_ecog.zip''' * Unzip it outside of any of the Brainstorm folders (program folder or database folder) * '''Brainstorm''': * Start Brainstorm (Matlab scripts or stand-alone version). * Select the menu File > Create new protocol. Name it "'''TutorialEcog'''" and select: <<BR>>"'''No, use individual anatomy'''", <<BR>>"'''No, use one channel file per acquisition run'''". |
| Line 31: | Line 28: |
| The pre-implantation T1 MRI is available as a .nii file (sample_ecog/sub-ecog01/ses-postimp/anat). This volume was processed with [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer|FreeSurfer 6]], and the output segmentation is available in the derivatives folder (sample_ecog/derivatives/freesurfer/sub-ecog01/ses-preimp). We don't need to import both to the Brainstorm database: in order to minimize the number of operations, we will import only the FreeSurfer folder. | The pre-implantation T1 MRI is available as a .nii file (sample_ecog/sub-ecog01/ses-postimp/anat). This volume was processed with [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer|FreeSurfer 7.1.1]], and the output segmentation is available in the derivatives folder (sample_ecog/derivatives/freesurfer/sub-ecog01_ses-preimp). We don't need to import both to the Brainstorm database: in order to minimize the number of operations, we will import only the FreeSurfer folder. |
| Line 36: | Line 33: |
| * Right-click on the subject node > '''Import anatomy folder''': * Set the file format: "FreeSurfer folder" * Select the folder: '''sample_ecog'''/'''derivatives'''/'''freesurfer'''/'''sub-ecog01'''/'''ses-preimp''' * Number of vertices of the cortex surface: 15000 (default value) * In the MRI Viewer, click on "[[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#MNI_transformation|Click here to compute MNI transformation]]". It computes an affine transformation between the subject space and the MNI ICBM152 space, and sets default positions for all the anatomical landmarks. * Click on [Save] to continue with the importation of the anatomy. At the end, make sure the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click to select it as the default cortex surface. <<BR>><<BR>> {{attachment:anatomy.gif||width="355",height="225"}} |
* Right-click on the subject node > '''Import anatomy folder (auto)''': [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Alternatives|More information]] * Set the file format: '''FreeSurfer + Volume atlases''' * Select the folder: '''sample_ecog'''/'''derivatives'''/'''freesurfer'''/'''sub-ecog01_''''''ses-preimp''' * At the end, make sure the file "cortex_15000V" is selected (downsampled pial surface), otherwise double-click to select it as the default cortex surface. <<BR>><<BR>> {{attachment:anatomy.gif}} * Rename the file '''MRI''' into '''T1pre''', for better readability in the rest of the tutorial. <<BR>><<BR>> {{attachment:mri_rename.gif}} * When processing your own datasets: Instead of using FreeSurfer, you can decide to run the segmentation of the pre-implantation MRI directly from Brainstorm, using [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]]. * For help with using the volume atlases, refer to the introduction tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Subcortical_regions:_Volume|Explore the anatomy]]. |
| Line 45: | Line 46: |
| * The pre-implantation MRI will be used as the anatomical reference for this subject. We will now import a second scan done after the sEEG/ECoG implantation, on which we can see the recording contacts. In this dataset, the post-implantation volume is another T1 MRI scan (contacts hyposignal appear in black), but it is more commonly a CT scan (contacts hypersignal appear in white). | * The pre-implantation MRI will be used as the anatomical reference for this subject. We will now import a second scan done after the sEEG/ECoG implantation, on which we can see the recording contacts. In this dataset, the post-implantation volume is another T1 MRI scan (contacts hyposignal appears in black), but it is more commonly a CT scan (contacts hypersignal appears in white). |
| Line 50: | Line 51: |
| * Right-click on the post-implantation volume > '''Register with default MRI > Reslice / vox2ras'''. <<BR>><<BR>> {{attachment:anat_reslice.gif}} * Right-click on the resliced post-implantation volume > <<BR>><<BR>> {{attachment:anat_overlay1.gif}} * The MRI viewer opens, showing the post-implantation volume as a colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section "Data options" of the Surface tab, adjust its [[Tutorials/Colormaps|colormap]] with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well. <<BR>><<BR>> {{attachment:anat_overlay2.gif||width="590",height="397"}} |
* Rename the new volume '''sub-ecog01_ses-postimp_T1w_spm''' into '''T1post_spm'''. <<BR>><<BR>> {{attachment:mri_rename2.gif}} * Right-click on the T1post volume > '''Register with default MRI > Reslice / world coordinates'''. <<BR>><<BR>> {{attachment:anat_reslice.gif}} * Right-click on the resliced T1post volume > Display > Overlay on default MRI<<BR>><<BR>> {{attachment:anat_overlay1.gif}} * The MRI viewer opens, showing the post-implantation volume as a colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section "Data options" of the Surface tab, adjust its [[Tutorials/Colormaps|colormap]] with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well. <<BR>><<BR>> {{attachment:anat_overlay2.gif}} |
| Line 55: | Line 57: |
| * Right-click on the subject folder > '''Generate BEM surfaces'''. Keep the default options. * When the computation is done, double-click on the the original head surface ("head mask") instead of the new BEM version, which is not estimated well because the volume was defaced. <<BR>><<BR>> {{attachment:anat_bem.gif||width="562",height="228"}} |
* Skull surfaces are useful for defining the position of ECOG contacts, and displaying them in 3D views. The contacts are projected on the inner skull surfaces, and then they are represented as cylinders oriented tangentially to the skull. * Right-click on the subject folder > '''Generate BEM surfaces > Brainstorm'''. Keep the default options. * When the computation is done, double-click on the the original head surface ("head mask") to select it as the default instead of the new BEM version, which is not estimated well because the volume was defaced. <<BR>><<BR>> {{attachment:anat_bem.gif}} <<BR>><<BR>> {{attachment:anat_bem2.gif}} |
| Line 72: | Line 75: |
| == Editing the contacts positions == For 3D mapping and to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the grids and depth electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. This tutorial will now explain how to do this manual marking with the MRI Viewer from the post-implantation MRI, which shows clear hyposignal artifacts around the ECoG and sEEG contacts. We prefer using the non-resliced volume because it offers a clearer view of these artifacts. ''If you have no interest in this or if you want to skip faster to the analysis of the signals, you can load the 3D positions from the BIDS dataset: right-click on the channel file > Add EEG positions > Import from file, select format "EEG: BIDS electrodes.tsv, subject space mm" and file sample_ecog/sub-ecog01/ses-postimp/ieeg/sub-ecog01_ses-postimp_space-other_electrodes.tsv, use transformation=Yes.'' * Right-click on the channel file > MRI registration > Edit (MRI Viewer: sub-ecog01_ses-postimp_T1w.nii_vox2ras_spm). * Adjust the brightness and contrast of the colormap in order to see the contacts artifacts correctly: right-click on the figure and move your mouse up/down, or left-click on the colormap and move your mouse up/down (brightness) or left/right (contrast), or right-click on the figure > Colormap:Anatomy. |
== Edit the contacts positions == For 3D mapping and to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the grids and sEEG electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. This tutorial will now explain how to do this manual marking with the MRI Viewer from the post-implantation MRI, which shows clear hyposignal artifacts around the ECoG and sEEG contacts. We prefer using the non-resliced volume because it offers a clearer view of these artifacts. If you have no interest in this or if you want to skip faster to the analysis of the signals, you can load the 3D positions from the BIDS dataset: read the advanced section [[https://neuroimage.usc.edu/brainstorm/Tutorials/ECoG#Alternative:_Read_BIDS_electrodes.tsv|Read BIDS electrodes.tsv]]. |
| Line 81: | Line 81: |
| * Right-click on the channel file > '''MRI registration''' > '''Edit (MRI Viewer: T1post_spm)'''. * Adjust the brightness and contrast of the colormap in order to see the contacts artifacts correctly: * right-click on the figure and move your mouse up/down, or * left-click on the colormap and move your mouse up/down (brightness) or left/right (contrast), or * right-click on the figure > Colormap:Anatomy (figures below: contrast=100, brightness=100) * For help with all the options iEEG tab, refer to the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Panel_iEEG|Epileptogenicity tutorial]].<<BR>>Related tutorial: [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Edit_implantation_without_recordings|Edit implantation without recordings]]. |
|
| Line 89: | Line 96: |
| * Navigate in the volume and position the cursor at the center of the artifact corresponding to contact '''G_A1''' (look at the implantation schemes above, it is the more frontal and mesial in the grid), then click on button ('''#1''') in the iEEG tab. <<BR>>If you can't find this point, you could enter manually the coordinates of the point: click on the button "Set current coordinates" at the bottom of the MRI Viewer window, and copy the MRI coordinates from the screen capture below. <<BR>><<BR>> {{attachment:edit_ga1.gif}} * Place the cursor at contact '''G_A8''' and click on button ('''#2''').<<BR>><<BR>> {{attachment:edit_ga8.gif}} * Place the cursor at contact '''G_H8''' and click on button ('''#3''').<<BR>><<BR>> {{attachment:edit_gh8.gif}} * Place the cursor at contact '''G_H1''' and click on button ('''#4''').<<BR>><<BR>> {{attachment:edit_gh1.gif}} * After placing the last corner, it should start computing for a while. The positions of the 64 contacts are distributed in the polygon you defined, and then projected to the surface of the brain. To get the figure below, with all the contacts projected on all slices, select the checkbox "MIP: Functional" in the MRI Viewer.<<BR>><<BR>> {{attachment:edit_g_done.gif}} * In this window, the contacts can be moved by simple drag-and-drop of the dots representing them. If you move them by mistake, there is unfortunately no undo button. But you can cancel your modifications in two ways: close the figure and discard the modifications, or select "G" in the iEEG tab and then use the menu '''Contacts > Use default positions''', to recompute the initial positions from the 4 four corners of the ECoG grid. In the same menu Contacts, you can find additional projection options: '''Project on inner skull''' and '''Project on cortex'''. All these menus work well only when skull surfaces are available for the patient (see the section above that explains how to use the menu '''Generate BEM surfaces'''). * After adding successfully an element, you can select menu '''Contacts > Save modifications''', to make sure you will not lose the work you've already done. |
* Navigate in the volume and position the cursor at the center of the artifact corresponding to contact '''G_A1''' (look at the implantation schemes above, it is the more frontal and mesial in the grid), then click on button '''[#1]''' in the iEEG tab. <<BR>>If you can't find this point, you could enter manually the coordinates of the point: click on the button "'''Set current coordinates'''" at the bottom of the MRI Viewer window, and copy the MRI coordinates from the screen capture below. <<BR>><<BR>> {{attachment:edit_ga1.png}} * Zoom in/out with the buttons or the associated shortcuts (Ctrl+Scroll or +/-) and explore the volume with (Shift+)x/y/z. Additional display options are available in the popup menu for this figure. All the shortcuts are listed in [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Using_the_MRI_Viewer|this tutorial]]. * Place the cursor at contact '''G_A8''' and click on button '''[#2]'''.<<BR>><<BR>> {{attachment:edit_ga8.gif}} * Place the cursor at contact '''G_H8''' and click on button '''[#3]'''.<<BR>><<BR>> {{attachment:edit_gh8.gif}} * Place the cursor at contact '''G_H1''' and click on button '''[#4]'''.<<BR>><<BR>> {{attachment:edit_gh1.gif}} * After placing the last corner, it should start computing for a while. The positions of the 64 contacts are distributed in the polygon you defined, and then adjusted to the shape of the brain. To get the figure below, with all the contacts projected on all slices, select the checkbox "'''MIP: Functional'''" in the MRI Viewer.<<BR>><<BR>> {{attachment:edit_g_done.gif}} * In this window, the contacts can be moved by simple '''drag-and-drop''' of the dots representing them. If you move them by mistake, there is unfortunately no undo button. But you can cancel your modifications in two ways: close the figure and discard the modifications, or select "G" in the iEEG tab and then use the menu '''Contacts > Use default positions''', to recompute the initial positions from the 4 four corners of the ECoG grid. * In the same menu Contacts, you can find additional projection options: '''Project on inner skull''' and '''Project on cortex'''. All these menus work well only when skull surfaces are available for the patient (see the section above that explains how to use the menu '''Generate BEM surfaces'''). * From time to time, you can click on the menu '''Contacts > Save modifications''' in the iEEG tab. This would prevent you from losing your work if anything unexpected happens. |
| Line 104: | Line 113: |
| * Contact diameter: '''4 mm <<BR>>''' * ECoG strips are defined only with two points, the first and the last contact. In the convention used here, the first contact is always the deepest one. |
* Contact diameter: '''4 mm''' * ECoG strips are defined only with two points, the first and the last contact. The intermediate contacts are uniformly distributed between the first and last point. In the convention used here, the first contact is always the deepest one. |
| Line 107: | Line 116: |
| * Place the cursor on '''IHA1''' (the deepest contact on the most frontal strip) and click ('''#1'''). <<BR>><<BR>> {{attachment:edit_iha1.gif}} * Place the cursor on '''IHA4''' and click ('''#2''').<<BR>><<BR>> {{attachment:edit_iha4.gif}} * Because you have selected the type '''ECOG-mid''', the contacts are not be projected on the skull or the cortex automatically. The contacts between the first and the last one are by default distributed uniformly on a straight line, which probably doesn't match their actual positions. Move slightly contacts IHA2 and IHA3 so that they appear in the middle of the artifacts. |
* Place the cursor on '''IHA1''' (the deepest contact on the most posterior strip) and click '''[#1]'''. To jump directly to the coordinates of this point: click on the button '''Set current coordinates''' at the bottom of the MRI viewer, and report the MRI coordinates displayed in the screen capture below.<<BR>><<BR>> {{attachment:edit_iha1.gif}} * Place the cursor on '''IHA4''' and click '''[#2]'''.<<BR>><<BR>> {{attachment:edit_iha4.gif}} * Because you have selected the type '''ECOG-mid''', the contacts are not be aligned on the skull or the cortex automatically. The contacts between the first and the last one are by default distributed uniformly on a straight line, which probably doesn't match their actual positions. Move slightly contacts IHA2 and IHA3 so that they appear in the middle of the artifacts. |
| Line 112: | Line 121: |
| * Repeat the same steps with '''IHB''': Set it as '''ECOG-mid''', contact distance=10mm. * Place the cursor on '''IHB1''' (the deepest contact on the middle strip) and click ('''#1''').<<BR>><<BR>> {{attachment:edit_ihb1.gif}} * Place the cursor on '''IHB4''' and click ('''#2''').<<BR>><<BR>> {{attachment:edit_ihb4.gif}} |
* Repeat the same steps with '''IHB''': Set it as '''ECOG-mid''', contact spacing='''10mm'''. * Place the cursor on '''IHB1''' (the deepest contact on the middle strip) and click '''[#1]'''.<<BR>><<BR>> {{attachment:edit_ihb1.gif}} * Place the cursor on '''IHB4''' and click '''[#2]'''.<<BR>><<BR>> {{attachment:edit_ihb4.gif}} |
| Line 119: | Line 128: |
| * Repeat the same steps with '''IHC''': Set it as '''ECOG-mid''', contact distance=10mm. * Place the cursor on '''IHC1''' (the deepest contact on the posterior strip) and click ('''#1''').<<BR>><<BR>>{{attachment:edit_ihc1.gif}} * Place the cursor on '''IHC4''' and click ('''#2''').<<BR>><<BR>> {{attachment:edit_ihc4.gif}} |
* Repeat the same steps with '''IHC''': Set it as '''ECOG-mid''', contact spacing='''10mm'''. * Place the cursor on '''IHC1''' (the deepest contact on the anterior strip) and click '''[#1]'''.<<BR>><<BR>> {{attachment:edit_ihc1.gif}} * Place the cursor on '''IHC4''' and click '''[#2]'''.<<BR>><<BR>> {{attachment:edit_ihc4.gif}} |
| Line 124: | Line 133: |
| === sEEG depth electrodes: TA, TB === * In the iEEG tab, click on button ['''Set tip'''], the button turns green. * In the MRI Viewer, place the cursor at the entrance of the electrode in the skull, or anywhere else along the depth electrode if this is more convenient. We don't use the actual position of this point, we only use it to estimate the direction of the electrode from its tip. Then click on button ['''Set skull entry''']. The button turns green and the electrode appears in the MRI viewer. * If there are contacts available for this electrode, this would also place them along the depth electrode. If the positions for these contacts was already defined, you should get a warning asking whether you want to update their positions. * From time to time, you can click on the menu '''Contacts > Save modifications''' in the iEEG tab. This would prevent you from losing your work if anything unexpected happens. * When you are done with marking all the electrodes, click on ['''Save'''] in the MRI viewer. You can then display the electrodes you placed in 3D (right-click on the channel file > Display sensors). |
=== sEEG electrodes: TA, TB === '''TA''' * In the iEEG tab, select electrode '''TA''' and edit the options: * Type: __'''SEEG'''__ * Number of contacts: '''10''' * Contact spacing: '''10 mm''' * Contact length: '''2.5 mm''' * Contact diameter: '''1 mm''' * Electrode diameter: '''0.9 mm''' * Electrode length: '''120 mm''' * The contacts actually have the same diameter as the electrode itself, but we use a slightly higher diameter for display, so that they render well in most 3D and 2D views. * sEEG electrodes are defined with two points, the tip of the electrode (deepest contact) and a second point anywhere along the axis the electrode. One typical solution is to use the entry point in the skull as this second point, because the screws are always easy to identify. In this example, the electrodes are curved, and the entry point is not aligned with most of the contact, therefore we suggest to pick the last contact on the electrode instead. The intermediate contacts are placed along electrode, separated according to the "contact spacing" option, starting from the tip. * Place the cursor on '''TA1''' (the deepest contact on the more mesial probe) and click '''[Set tip]'''. To jump directly to the coordinates of this point: click on the button '''Set current coordinates''' at the bottom of the MRI viewer, and report the MRI coordinates displayed in the screen capture below.<<BR>><<BR>> {{attachment:edit_ta_tip.gif}} * Place the cursor on '''TA10''' (the last contact on this electrode) and click '''[Set skull entry]'''. <<BR>>This is obviously not the location of the screw in the skull, but as the electrode is bent, this is a better point to approximate the contact locations on a straight line.<<BR>><<BR>> {{attachment:edit_ta_entry.gif}} * The electrode is curved, therefore you need to move most of the intermediate contacts '''TA2-TA9''' to more appropriate positions, at the center of the matching artifact. Unfortunately, Brainstorm does not handle well curved electrodes yet: if you move the contacts, the electrode remains represented as a straight line. This is a display issue only, which has no impact on any computation, but we hope to improve it at some point. '''TB''' * Repeat the same steps with '''TB'''. with the same configuration: * Type: __'''SEEG'''__ * Number of contacts: '''10''' * Contact spacing: '''10 mm''' * Contact length: '''2.5 mm''' * Contact diameter: '''1 mm''' * Electrode diameter: '''0.9 mm''' * Electrode length: '''120 mm''' * Place the cursor on '''TB1''' (the deepest contact on the more mesial probe) and click '''[Set tip]'''.<<BR>><<BR>> {{attachment:edit_tb_tip.gif}} * Place the cursor on '''TB10''' (the last contact on this electrode) and click '''[Set skull entry]'''. <<BR>><<BR>> {{attachment:edit_tb_entry.gif}} * Move the intermediate contacts '''TB2-TB9''' to the centers of the artifacts. * When you are done with marking all the electrodes, click on ['''Save'''] in the MRI viewer. <<BR>><<BR>> {{attachment:edit_done.gif||width="545",height="396"}} === 3D display === * Right-click on the channel file > '''Display sensors''' > Explore all the options available. <<BR>>For help with the display options in the iEEG tab: [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Panel_iEEG|Epileptogenicity tutorial]].<<BR>>You can add more anatomy elements to the figure with the button "Add a surface" at the top-right of the Surface tab. For more help: [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#MRI_in_3D|Display the anatomy]]. <<BR>><<BR>> {{attachment:display_menu.gif}} {{attachment:display_3d.gif||width="631",height="202"}} === Project on brain surface === You probably noticed that many the contacts of the ECoG grid appear under the surface of the cortex in these 3D figures. This is mostly due to the '''brain shift''' between the volumes we used to mark the contacts (post-implantation MRI) and the volume from which the cortex surface was reconstructed (pre-implantation MRI). One solution to compensate for this brain shift and produce more readable figures is to project the contacts on the pre-implantation surfaces. * In the iEEG tab, select the grid '''G''', then use menu '''Contacts > Project on inner skull'''.<<BR>><<BR>> {{attachment:display_project.gif||width="627",height="247"}} * You could also test what you obtain by projecting the contacts on the '''inner skull'''. It produces similar results, but with a different method, projecting the contacts possibily a bit further away from the brain. Depending on the quality of the surfaces, the shape of the brain, and the position of the contacts, one solution may produce weird results, try the other one in that case. |
| Line 134: | Line 178: |
| === Edit implantation without recordings === You can also start defining an implantation scheme without any recordings in your database. You can use this option for creating a text file with all the contacts positions, and use it in Brainstorm or any other program. * Go back to the subject anatomy, right-click on the appropriate post-implantation volume > '''SEEG/ECOG implantation''' * It creates a new folder "Implantation" in the subject, and starts editing an empty channel file. You need to create the electrodes and set their models (or all their properties) before placing them in the MRI viewer. When you place an electrode, it creates all the corresponding contacts, while in the previous case, it was only setting the position of the existing contacts. * At any moment, you can export the 3D positions of the contacts you created to a text file with the menu '''Contacts > Export contacts positions '''in the iEEG tab. * When you are done, click on [Save] in the MRI viewer. It saves the new channel file and updates the number of channels in the database explorer. == Display the depth electrodes == === 3D figures === * Right-click on the channel file > '''Display sensors''' > Explore all the options available. <<BR>><<BR>> {{attachment:display_menu.gif}} * You can render the SEEG depth electrodes in 3D together with the subject anatomy: surfaces, pre- or post-implantation volumes. You can add more anatomy elements to the figure with the button "Add a surface" at the top-right of the Surface tab. For more help: [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#MRI_in_3D|Display the anatomy]]. <<BR>><<BR>> * Click on a contact to select it, right-click on it to get its name: <<BR>><<BR>> === MRI Viewer === * You can also display the contacts in the MRI viewer, on top of the the pre- or post-implantation volumes. By default, the electrode is displayed in a slice if there is a SEEG contact associated to it in the slice.<<BR>><<BR>> * To display all the electrodes, select the option "'''MIP: Functional'''". For a glass-brain view, select at the same time the option "'''MIP: Anatomy'''". <<BR>><<BR>> * Zoom in/out with the buttons or the associated shortcuts (Ctrl+Scroll or +/-) and explore the volume with (Shift+)x/y/z. Additional display options are available in the popup menu for this figure. All the shortcuts are listed in [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Using_the_MRI_Viewer|this tutorial]]. * You can '''edit the position of a contact''' directly by clicking on its dot in one of the slice and moving it around. Modifications are not saved immediately and can be cancelled when you close the window. Alternatively, you can right-click on the figure > '''Set electrode position'''. === Panel iEEG === * When opening SEEG/ECOG recordings, the panel iEEG is added to the Brainstorm window. You can use it to edit the display properties of the depth electrodes. In this interface, "'''electrode'''" refers to entire depth electrode implanted in the head of the patient while "'''contact'''" refers to recording sites on the electrode. There are multiple contacts on an electrode, and one contact corresponds to one channel of data in the channel file and the recordings. <<BR>><<BR>> {{attachment:panel_ieeg.gif||width="611",height="319"}} * SEEG depth electrodes are graphical objects, they are defined independently from the SEEG contacts available in the channel file and recordings. A contact/channel is associated to a depth electrode using the '''Group''' property of the channel, accessible with a right-click on the channel file > '''Edit channel file'''. * In this example, the depth electrodes available in the panel iEEG have been detected automatically based on the names and positions of the data channels. In the convention used here, the contact names must start with one or more letters (the name of the electrode) followed by a number representing the index of the contact on the electrode. Contact #1 is at the tip of the electrode, and is therefore the deeper contact of the electrode. If the convention used in your recordings is different, you may have to edit the electrodes properties in order to get them displayed correctly. * Buttons in the toolbar: * '''Add an electrode''': Adds an entry for a new depth electrode. The new electrode will not be displayed until you set its properties and position. This will not add or remove SEEG contacts or channels of data. * '''Remove selected electrodes''': Deletes a depth electrode from the list, but does not modify the list of SEEG contacts or channels of data. * '''Set color for selected electrodes''': Self explanatory. * '''Show/hide selected electrodes''': To hide an electrode in the 3D figures and MRI viewer, select it in the list then click on this button. Select all the electrodes with the standard shortcut Ctrl+A. * '''Display contacts as''': Depth electrodes / Spheres. * '''Contacts > Set default positions''': For each of the selected electrodes, the current positions of the SEEG contacts are discarded and replaced with the default positions of the contacts on the electrode. The properties used for setting the position of the contacts are the contact spacing, the tip of the electrode and the entry point in the skull. Contact #i is placed along the electrode at (i-1)*contact_spacing millimeters from the tip of the electrode. * '''Contacts > Project on electrode''': For each of the selected electrodes, the contacts are projected orthogonally on the electrode. This menu can be useful for aligning contacts that were marked one by one. * '''Contacts > Save modifications''': Save the current modifications to the channel file. Otherwise, the modifications are saved only when you close the figure (dialog box "Save modifications to channel file?") * '''Contacts > Export contacts positions''': Save the 3D positions of the SEEG contacts in a text file, using one of the file formats supported by Brainstorm. * Electrode properties: ''Properties that have an impact on the position of the contacts''.<<BR>> * If you edit the properties, the modifications will apply to all the selected electrodes in the current channel file. Check what is selected before making changes. * '''Type''': SEEG/ECOG * '''Model''': List of electrode models. If you select an entry in this menu, it will copy the default properties for this model to the selected electrodes. If you are using electrodes that are not in this list, please post on the specification of your devices on the user forum and we will add them to this list. * '''Number of contacts''': Number of recording sites on the electrode. By default, this is set for SEEG to the maximum index found in a group of contacts. Example: electrode s' is associated to channels s'1, s'2, s'3, s'10, s'11, s'12 => detected number of contacts is 12. * '''Contact spacing''': Distance between the centers of two consecutive contacts in the electrode. In this example dataset, the default value corresponds to the average distance observed between pairs of adjacent contacts. * '''Set tip''': Select one electrode in the list, then move the cursor of the MRI viewer to the tip of the selected electrode (center of the first contact), and finally click on [Set tip]. * '''Set skull entry''': Select one electrode in the list, then move the cursor of the MRI viewer to the point where the depth electrode enters the skull, and finally click on [Set skull entry]. This position does not correspond to any contact, it is used only to estimate the direction of the depth electrode. [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Editing_the_contacts_positions|More details]]. * Display options: Properties that only affect the way the electrodes are renderer graphically. * '''Contact length''': Defines the length of the yellow cylinder that represents the contacts along the electrode axis, or the diameter of the sphere when the electrodes are not rendered. * '''Contact diameter''': Diameter of the yellow cylinders representing the contacts. By default, this value is slightly larger than the electrode diameter so that it is rendered correctly. If you use the same value as the electrode diameter, the contacts might not be visible. * '''Electrode diameter''': Diameter of the cylinder representing the SEEG depth electrode. * '''Electrode length''': Length of the cylinder representing the SEEG depth electrode. == Display the SEEG recordings == === SEEG time series === * Right-click on any "Link to raw file" > '''SEEG '''> '''Display time series'''.<<BR>><<BR>> {{attachment:display_ts.gif||width="637",height="276"}} |
== Alternative: Read BIDS electrodes.tsv == A BIDS dataset may contain information about the localization of the iEEG contacts in different systems of coordinates. These files are documented in the in [[https://bids-specification.readthedocs.io/en/stable/04-modality-specific-files/04-intracranial-electroencephalography.html#electrode-description-_electrodestsv|BIDS specification]]. In this dataset, we can find the following files in the folder ''sample_ecog/sub-ecog01/ses-postimp/ieeg''. Files * sub-ecog01_ses-postimp_'''space-IXI549Space'''_'''coordsystem.json''': MNI space metadata * sub-ecog01_ses-postimp_'''space-IXI549Space'''_'''electrodes.tsv''': XYZ coordinates, MNI space * sub-ecog01_ses-postimp_'''space-ScanRAS'''_'''coordsystem.json''': Subject space metadata * sub-ecog01_ses-postimp_'''space-ScanRAS'''_'''electrodes.tsv''': XYZ coordinates, subject space The tag '''space-''' indicates how the coordinates should be interpreted ([[https://bids-specification.readthedocs.io/en/stable/99-appendices/08-coordinate-systems.html|list of possible values]]): * '''IXI549Space''': Refers to a MNI normalization computed by SPM12 (see [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|Coordinate systems]]). You should be able to load these locations on any volume for which MNI coordinates are available, but with the approximations due to the volume-based normalization and possible errors due to the use of different types of normalization. In this particular dataset, the MNI coordinates were computed from Brainstorm with the [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#Non-linear_normalization|SPM12 Segment]] non-linear normalization. When possible, always prefer importing coordinates in subject space. * '''ScanRAS''': Refers to the scanner coordinates (named [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#World_coordinates|world coordinates]] in Brainstorm) of a specific anatomical image (MRI or CT). The interpretation of the coordinates in the file '''electrodes.tsv''' requires the information available in the adjacent file '''coordsystem.json'''. Typically, this pair of files defines coordinates associated to a specific volume in the BIDS dataset. For the subject space (space-ScanRAS), the contents of the json file is the following: {{{ "iEEGCoordinateSystem": "ScanRAS", "iEEGCoordinateUnits": "mm", "iEEGCoordinateSystemDescription": "Scanner-based RAS coordinates of the post-implantation T1 MRI", "iEEGCoordinateProcessingDescription": "SEEG contacts manually placed with Brainstorm on the post-implanation T1 MRI", "iEEGCoordinateProcessingReference": "https://neuroimage.usc.edu/brainstorm/Tutorials/ECoG", "IntendedFor": "sub-ecog01/ses-postimp/anat/sub-ecog01_ses-postimp_T1w.nii.gz" }}} The field '''IntendedFor''' points at the file ''sub-ecog01_ses-postimp_T1w.nii.gz''. This means that the coordinates in the files ''sub-ecog01_ses-postimp_space-ScanRAS_electrodes.tsv'' refer to the post-implantation T1 MRI. To add the coordinates in subject space (be careful: if you already marked the contacts manually, you would lose all your work - if you want to try this, duplicate the subject first): * Right-click on the channel file > '''Add EEG positions''' > '''Import from file''' * Select format "'''EEG: BIDS electrodes.tsv, subject space mm'''" <<BR>>(or adjust depending on the tsv file you select and the information defined in the json file) * Select file '''sub-ecog01_ses-postimp_space-ScanRAS_electrodes.tsv'''<<BR>><<BR>> {{attachment:import_tsv.gif}} Note that the electrodes.tsv file does not include all the information we defined manually in the previous section (ECOG vs. ECOG-mid, contact diameter...), therefore for optimal display you might need to go through the configuration of each implanted device in the iEEG tab and edit the values as previously. <<TAG(Advanced)>> == Display the recordings == === ECoG/sEEG time series === * Right-click on any "Link to raw file" > '''ECOG+SEEG '''> '''Display time series'''.<<BR>><<BR>> {{attachment:review_ts.gif||width="701",height="341"}} |
| Line 191: | Line 222: |
| * Change the montage (bipolar/monopolar): Record tab > Drop-down menu.<<BR>>Note all the default montages available for SEEG: you can display all the contacts at once, or each electrode individually. | * Change the montage (bipolar/monopolar): Record tab > Drop-down menu. |
| Line 196: | Line 227: |
| * With the menus "'''3D electrodes'''" you can visualize the SEEG values at the current time point. Move the time cursor and it will update the values represented on the SEEG contacts. <<BR>><<BR>> {{attachment:display_3delectrodes.gif||width="640",height="299"}} | * Montage options include, for each group separately or all the channels together: * '''Bipolar 1''': Difference of pairs of adjacent sensors (sparse), ''eg. A1-A2, A3-A4, A5-A6'' * '''Bipolar 2''': Difference of pairs of adjacent sensors (exhaustive), ''eg. A1-A2, A2-A3, A3-A4'' * '''Local average reference''': Average reference computed independently for each group === 2D/3D topography === With the menus [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreRecordings#A2D_Layout|2DLayout]]''', 2D Electrodes''' and '''3D Electrodes''' you can visualize the ECoG/sEEG values at the current time point. Move the time cursor and it will update the values represented on the contacts. {{attachment:review_topo.gif||width="725",height="272"}} |
| Line 199: | Line 238: |
| * From the same SEEG display menu, you can also project the SEEG values on the MRI or the cortex. You can edit the colormap for the displayed values [[http://neuroimage.usc.edu/brainstorm/Tutorials/Colormaps|like any other colormap]]. * '''MRI volume''': All the voxels in the neighborhood of a contact are attributed the value associated with this contact. In the figure below, both "MIP: functional" and "MIP: anatomy" options are selected (maximum intensity projection along each axis = "glass brain" view). * '''Cortex surface''': The values of the vertices are interpolated from the nearest SEEG contacts (the magnitude decreases with the distance to the contact). <<BR>><<BR>> {{attachment:display_interp_anat.gif||width="640",height="263"}} === 2D topography === Two additional popup menus allow you to display the SEEG recordings grouped by electrode, but without any 3D information: [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreRecordings#A2D_Layout|2DLayout]] and '''2DElectrodes'''. {{attachment:display_2dlayout.gif||width="705",height="199"}} == Review recordings == |
From the same ECOG+SEEG display menu, you can also project the recordings on the MRI or the cortex. You can edit the colormap for the displayed values [[http://neuroimage.usc.edu/brainstorm/Tutorials/Colormaps|like any other colormap]]. '''MRI volume''': All the voxels in the neighborhood of a contact are attributed the value associated with this contact. In the figure below, both "MIP: functional" and "MIP: anatomy" options are selected (maximum intensity projection along each axis = "glass brain" view). '''Cortex surface''': The values of the vertices are interpolated from the nearest SEEG contacts (the magnitude decreases with the distance to the contact). {{attachment:review_interp.gif}} |
| Line 212: | Line 249: |
| * In Process1, select all the continuous files (SZ1, SZ2, SZ3: link to raw files or folders). * Run process '''Frequency''' > '''Power spectrum density (Welch)''': All file, Length=10s, Overlap=50%. <<BR>><<BR>> {{attachment:psd_process.gif||width="611",height="246"}} * Double-click on the PSD files to display them. Some channels have noise levels that are obviously higher than the others and should therefore be considered as suspicious. <<BR>>Left = SZ1 (v'1,f'1), Center = SZ2 (v'1,t'8), Right = SZ3 (o'1,t'8)<<BR>><<BR>> {{attachment:psd_result.gif||width="614",height="294"}} |
* In Process1, select the continuous file. * Run process '''Frequency''' > '''Power spectrum density (Welch)''': All file, Length=10s, '''ECOG,SEEG'''. * Double-click on the PSD files to display them, change to a log-scale for the X axis, zoom in. None of the channels seem to have noise levels that are higher or lower than the others, all the channels are possibly good. <<BR>><<BR>> {{attachment:review_psd.gif||width="702",height="224"}} |
| Line 217: | Line 254: |
| === Bad channels === We will now to review the recordings of the three seizures recorded for this subject: SZ1, SZ2, SZ3. All the following steps are illustrated only for '''SZ1''' but need to be reproduced for the other files. * Double-click on '''SZ1/Link to raw file''' to open the SEEG recordings for the first seizure. * We need to mark the bad channels using a "'''monopolar'''" montage (common reference). If you are currently reviewing the recordings with a bipolar montage, switch it back to "All channels", using the menu in the toolbar of the record tab or with the popup menu of the time series figure. Otherwise, marking a signal as bad (eg. "v'2-v'1") would actually mark two channels as bad ("v'1" and "v'2"), while only one contact ("v'1") is not recording properly. * Click on the channels that you consider as bad to select them. For SZ1, select '''v'1''' and '''f'1'''. * Press the '''delete''' key, or use the popup menu Channels > '''Mark selected as bad'''. <<BR>><<BR>> {{attachment:mark_bad.gif||width="465",height="217"}} * Close the figure and repeat for the two other files. |
|
| Line 227: | Line 255: |
| At this point of the analysis, we are still looking at the original files, no SEEG data was copied to the database. The montages are saved in the Brainstorm preferences, the bad channels and new events are saved in the links of the database but not reported to the original .TRC files. If you delete your protocol at this point, you would only lose the event marking and bad channel selection. We are now going to import two segments of recordings for each seizure file: the seizure (10s before and 40s after the onset) and the baseline (all the segment selected). This will make real copies of the data in the database, so we can run additional processes on them. === Import in database === * Right-click on '''SZ1 / Link to raw file''' > '''Import in database''': * Time window: All, the only interesting point is to have access to the Onset marker. |
At this point of the analysis, we are still looking at the original files, no data was copied to the database. The montages are saved in the Brainstorm preferences, the bad channels and new events are saved in the links of the database but not reported to the original files. If you delete your protocol at this point, you would only lose the event marking and bad channel selection. We are now going to import a segment a recordings around each of the three seizures marked in this file (10s before and 20s after the onset). This will make real copies of the data in the database, so we can run additional processes on them. * Double-click on the on the "Link to raw file" to open the time series figure (ECOG+SEEG). * In the Record tab: double-click on the event category '''seizure_begin''' and rename it '''seizure'''. * Close the file viewer to save the modification to the events. * Right-click on '''Link to raw file''' > '''Import in database''': * Time window: All |
| Line 235: | Line 265: |
| * Use events: Enabled, select '''Onset''' * Epoch time: '''[-10000, +40000] ms''' (imports -10s to 40s around the event "Onset") |
* Use events: Enabled, select '''seizure''' * Epoch time: '''[-10000, +20000] ms''' (imports -10s to 20s around the event "seizure") |
| Line 239: | Line 269: |
| * Create a separate folder for each event type: Disabled <<BR>><<BR>> {{attachment:import_onset.gif||width="601",height="262"}} | * Create a separate folder for each event type: Disabled <<BR>><<BR>> {{attachment:import_seizures.gif||width="702",height="245"}} |
| Line 241: | Line 271: |
| * The imported epochs are saved with a new timing: for the seizure onsets, the reference time t=0s is now the event "Onset", which has been removed from the list. You can still see the other marker Seizure, which should be slightly before 0s. For the baselines, the reference t=0 is the beginning of the segment.<<BR>><<BR>> {{attachment:import_database.gif||width="632",height="283"}} == Time-frequency analysis (pre-onset baseline) == The aim of this section is to evaluate the frequency range for the computation of the epileptogenicity maps. We need to identify which frequency band is the most representation and specific of the high-frequency activity at the beginning of the seizures. Starting from this step, you need to have the [[http://www.fil.ion.ucl.ac.uk/spm/software/download/|SPM12 toolbox]] installed on your computer and added to your Matlab path. We need here the multitaper functions from the FieldTrip toolbox, which are also already in the SPM package. * In Process1, select all the Onset bipolar files. * Select the process: '''Frequency > FieldTrip: ft_mtmconvol (Multitaper)''': (do not run) * '''Time window''': [-10, 10]s * '''Sensor types''': SEEG * '''Taper''': hanning * '''Frequencies''': 10:3:220 * '''Modulation factor''': 10 * '''Time resolution''': 1000ms * '''Measure''': Magnitude * '''Save average''': Disabled * Add the process: '''Standardize > Baseline normalization''': * '''Baseline''': [-10, -1]s * '''Method''': Z-score transformation * '''Overwrite''': Enabled * Add the process: '''Average > Average files''': * '''Group files''': Everything * '''Function''': Arithmetic average * Run the pipeline.<<BR>><<BR>> {{attachment:timefreq_multitaper.gif||width="511",height="442"}} <<BR>><<BR>> {{attachment:timefreq_normalize.gif||width="630",height="496"}} * Four new files are available in the database: one for each bipolar Onset epoch, and one average, saved in folder "Intra-subject". * Right-click on the average time-frequency map > '''All channels'''. Then you can click on a channel to open it in a separate figure. Change the colormap to "GIN" and the colormap maximum to [-0.1,+0.1]*10^2 to reproduce the figures below. <<BR>><<BR>> == Time-frequency analysis (separate baseline) == Another approach is to normalize the time-frequency maps based on the baseline file used for the computation of the epileptogenicity maps, instead of the short baseline immediately before the seizure onset. This is a bit more complicated but produces maps that are more coherent with the epileptogenicity measures used in the next section. To increase the educational value of this section, let's manage the time differently: we'll compute the time-frequency decomposition for the entire Onset epochs (-10s,+40s) and then extract only the section we are interested in. * Start by deleting all the time-frequency files computed in the previous section: select them in the database explorer and press the Delete key, or select the subject folder in Process1 with button [Process time-freq] selected and use process File > Delete files > Delete selected files. * In Process1, select all the bipolar files (Onset and Baseline). * Run the process: '''Frequency > FieldTrip: ft_mtmconvol (Multitaper)''': * '''Time window''': All file * '''Sensor types''': SEEG * '''Taper''': hanning * '''Frequencies''': 10:3:220 * '''Modulation factor''': 10 * '''Time resolution''': 1000ms * '''Measure''': Magnitude * '''Save average''': Disabled <<BR>><<BR>> {{attachment:timefreq_adv_multitaper.gif||width="537",height="389"}} * In Process2, select the time-frequency file for the baseline in FilesA, and for the onset in FilesB. You can either select the time-domain epochs and click on the buttons [Process time-freq], or select directly the time-frequency files generated with the previous step. Note the order in these lists matters: make sure you select the Onset and Baseline files in the exact same order. * Select the process: '''Standardize > Baseline normalization''' (do not run immediately): * '''Baseline''': All file * '''Method''': Z-score transformation<<BR>><<BR>> * Add process: '''Average > Average files''': * '''Group files''': Everything * '''Function''': Arithmetic average <<BR>><<BR>> * Run the pipeline. In output, you get the three normalized time-frequency maps for the three Onset epochs, plus the average of these three files. As expected, the time definition of the average file is [-10,+40]s. In order to produce similar figures as the ones from the previous section, we will extract the same time window as before (-10s to +10s).<<BR>><<BR>> {{attachment:timefreq_adv_full.gif||width="525",height="165"}} * In Process1, select the average time-frequency map. * Run process '''Extract > Extract time''', time window = '''[-9.5, 9.53]s''' (FieldTrip's multitaper function cuts a piece of the recordings to account for possible edge effects). <<BR>><<BR>> === Create a movie with the SEEG signals === Saving the conclusions of your visual exploration as a video file is an efficient solution for keeping track of your analyses and sharing them with your colleagues. In such a movie, it can be interesting to display simultaneously the original SEEG recordings with the epileptogenicity maps. However, if you try to open the two files, you get an error message explaining that Brainstorm cannot display two files with different time definitions at the same time. A solution to go around this limitation is to resample the epileptogenicity results (one value every two seconds = 0.5Hz) with the same frequency as the SEEG recordings (initially 512Hz). This would multiply the size of the file containing the epileptogenicity results by 2*512, creating a file of several Gb. To avoid wasting too much disk space and risking to crash Matlab by creating gigantic variables, we can cut and downsample the SEEG recordings of interest. Let's illustrate this with the seizure SZ2. * In Process1, select the file '''SZ2_bipolar_2/Onset'''. * Select process '''Extract > Extract time''': 0-20 s * Add process '''Pre-process > Resample''': 128 Hz, Overwrite<<BR>><<BR>> {{attachment:movie_extract.gif||width="569",height="247"}} * Run the execution. The output is a downsampled version of the first 20s after the seizure onset. * In Process2, select: <<BR>>FilesA = Resampled seizure recordings '''SZ2_bipolar_2/Onset | time''' (target time definition)<<BR>>FilesB = Stat results '''Epileptogenicity_surface/EI_SZ2_120_200_3''' (file to resample) * Run process '''Standardize > Interpolate time''': You can try the interpolation methods "nearest" or "linear", depending on if you would like to see the real statistical results or smooth (but inaccurate) transitions between the steps of two seconds for which the epileptogenicity maps were computed. * The new resampled epileptogenicity file has the same time definition as the resampled seizure SEEG recordings: the two can now be opened simultaneously. * After creating all the files you want to display, open them simultaneously and position the various figures on your screen the way you would like them to appear in the movie. Note that the text is displayed with a fixed size, it doesn't change if you reduce the size of the figures, so if you make the figures smaller the text will be bigger relatively to the graphics, making it possibly more readable in your screen captures. * To display only a subset of channels in the figure, either use an existing predefined montage, create a montage with the Montage editor, or select the channels you want in the time series figures and use the menu "Create from selection".<<BR>><<BR>> * Right-click on any of the figures > Snapshot > '''Movie (time): All figures'''. You can experiment the effect of the various parameters to obtain exactly the movie you want. The figure on which you right-click will show a time stamp at the bottom-left corner in the movie. <<BR>><<BR>> * The video file generated can be viewed on any computed, added to a website or included in a slide of a presentation. <<BR>><<BR>> <<HTML(<center><iframe width="560" height="315" src="https://www.youtube.com/embed/1jpmc1Sl6OE" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe></center>)>> |
* The imported epochs are saved with a new timing: for the seizure onsets, the reference time t=0s is now the event "Onset", which has been removed from the list. You can still see the other marker Seizure, which should be slightly before 0s. For the baselines, the reference t=0 is the beginning of the segment.<<BR>><<BR>> {{attachment:display_epochs.gif}} == Anatomical labelling == As illustrated in the introduction tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Subcortical_regions:_Volume|Explore the anatomy]], all the anatomical atlases available in the subject (e.g. DKT, Desikan-Killiany, Destrieux) are available in the MRI Viewer. Select the current atlas from the figure popup menu, and the anatomical label under the cursor is displayed at the top-right corner. You can overlay all the labels on the MRI with the menu '''Show atlas'''. {{attachment:volatlas.gif}} We can use these anatomical parcellations, and also the surface parcellations of the cortex available as scouts, in order to label automatically all the ECOG/SEEG contacts. If accurate MNI coordinates are expected, it is recommended to use a [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#Non-linear_normalization|non-linear MNI normalization]] instead of the initial linear registration computed at the beginning of this tutorial, as illustrated in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Anatomical_labelling|SEEG epileptogenicity tutorial]]. In the anatomy view, right-click on the pre-implantation MRI (first file in the subject anatomy) > '''MNI normalization''' > '''Segment'''. This requires SPM12. Switch to the functional view. Right-click on the channel file > '''iEEG atlas labels''' > Select all the available options: XYZ coordinates in various [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems|coordinate systems]], volume parcellations, surface parcellations.<<BR>><<BR>> {{attachment:labelling1.gif}} When using surface atlases, select preferencially the high-resolution surfaces (~300000V): a higher density of labelled points on the surface will increase the chances to label correctly the contacts. Selecting the type of surface ("white", "mid" or "cortex") depends on multiple factors, and we don't have enough experience yet to give clear recommendations. To help you decide, here is what these labels mean: * '''White''': Represents the interface between the white matter and the gray matter * '''Cortex''': Inflated from the white surface, represents the interface between the gray matter and the CSF. * '''Mid''': In the FreeSurfer case, this surface is computed by Brainstorm as the layer half-way between the '''white''' and the '''cortex''' surfaces. If this is available, this might be your best guess. Try it yourself and please share your results and comments on the user forum. {{attachment:labelling2.gif}} {{attachment:labelling3.gif}} More options and details in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Anatomical_labelling|SEEG epileptogenicity maps]]. |
| Line 316: | Line 298: |
| == Video-EEG == <<TAG(Advanced)>> == Importing realistic surfaces == Instead of using the default SPM canonical surfaces, you can use realistic cortex envelopes, extracted with [[http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer|FreeSurfer]], [[http://neuroimage.usc.edu/brainstorm/Tutorials/SegBrainSuite|BrainSuite]] or [[http://neuroimage.usc.edu/brainstorm/Tutorials/SegBrainVisa|BrainVISA]]. Instead of importing the anatomy as described previously (pre MRI then post MRI), follow these steps. Note that this cannot be done with the simplified interface in the Guidelines tab. * Right-click on the subject folder > '''Import anatomy folder''': * Select file type: "BrainVISA folder" * Select folder: tutorial_epimap/anat/MRI/brainvisa/ * Number of vertices: 15000 * When the MRI viewer shows up asking for fiducial points, click on "'''Click here to compute MNI transformation'''", then click on [Save] when the computation is done. <<BR>><<BR>> {{attachment:anat_brainvisa.gif||width="361",height="194"}} * This is the anatomy of the subject (MRI pre-implantation) processed with the Morphologist pipeline of BrainVISA 4.5. The MRI pre-implantation is now imported, but the post-implantation is still missing. * Right-click on the subject folder > '''Import MRI''': * Import the volume exactly as previously, following [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Post-implantation_MRI|these guidelines]]. * When importing the volumes processed with FreeSurfer, you probably need to register the volume, as FreeSurfer rewrites the volume with a different orientation. Select option [SPM] instead of [Ignore]. * In the subject anatomy folder, you can decide to use the pial surface ("cortex") or the grey-white interface ("white") for the computation of the epileptogenicity maps. As these surfaces are fairly different, it may leed to very different results. A SEEG contact close to one surface can be too far from the other to be taken into account in the computation. Make this choice carefully. To change the default surface, right-click > '''Set as default cortex''', or simply double-click on it. Just avoid selecting the high-resolution surfaces (126000 vertices) or the computation will take forever. * Then you can repeat all the steps previously presented. The results may look different depending on the surface you selected (left=cortex, right=white). <<BR>><<BR>> {{attachment:anat_brainvisa_delay.gif||width="310",height="180"}} <<TAG(Advanced)>> == Volume coregistration == When importing two volumes successively in the subject anatomy, you need to coregister all the new volumes with the first volume imported, otherwise you wouldn't be able to do anything with them. The questions that are asked when importing a second volume are the following. * '''Transformation''': This has nothing to do with the coregistration with the first volume, you also got this question when importing the first volume. If you answer "yes" for the first volume, you should probably answer "yes" here as well.<<BR>><<BR>> {{attachment:coreg_transf.gif}} * '''Registration''': How to register the new volume with the previous one?<<BR>><<BR>> {{attachment:coreg_spm.gif}} * '''SPM''': Uses the SPM toolbox to estimate the rigid transformation from the new volume to the reference volume (the first volume imported). It modifies the voxel=>world transformation in the second volume (vox2ras 4x4 matrix). If you skip this registration step, you can run it later by right-clicking on the second file > '''Register with default MRI > SPM: Register only'''. * '''MNI''': Uses SPM code (embedded in Brainstorm) to estimate an affine transformation from each of the two volumes to the MNI normalized space. The transformation from the new volume to the reference is approximated with the two successive MNI transformations: (voxel_new=>MNI) => inv(voxel_ref=>MNI). This gives inaccurate results, and does not guarantee a rigid transformation (there might be slight scaling factor introduced). Do not use this option, unless you have no other option available. * '''Ignore''': Considers that the two volumes are already coregistered. This option will keep the original voxel=>world transformations available in the .nii files, and assume they bring the two volumes in the same space. * '''Reslice''': You have the option to rewrite the new volume so that it matches voxel by voxel the reference volume. If you know that the new file was already registered and resliced before you imported it in Brainstorm, answer NO to this question. Otherwise, this option is necessary if you are expecting to overlay the two volumes in the MRI viewer. If you do not reslice the volume, you would still be able to edit the positions of the SEEG contacts on one or the other volume, but not to superimpose them to check they were registered correctly. This is a limitation of the MRI viewer that will be improved at some point.<<BR>><<BR>> {{attachment:coreg_reslice.gif}} * All these options are also available after importing the file. Right-click on the second file > '''Register with default MRI'''. The reslice option previously presented corresponds to the menu "'''Reslice / vox2ras transform'''". The other reslicing options offer alternatives based on the MNI coordinates or the SCS coordinates, which are most of the time a lot less accurate. <<BR>><<BR>> {{attachment:coreg_popup.gif}} <<TAG(Advanced)>> == On the hard drive == The definition of the depth electrodes is saved in the channel file, in the field '''IntraElectrodes'''. It contains all the information that can be edited in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Panel_iEEG|iEEG tab]]. The position of the SEEG contacts is saved in the field Channel(i).Loc, as described in the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#On_the_hard_drive|Channel file]]. ChannelMat.IntraElectrodes is an array of structures with the following fields. * '''Name''': String identifying the depth electrode in the iEEG tab. * '''Type''': String identifying the type of device, 'SEEG' or 'ECOG'. * '''Model''': String identifying the make and model of the electrode (eg. 'DIXI D08-05AM Microdeep'). This is of known electrode models is accessible with panel_ieeg('GetElectrodeModels'). * '''Loc''': [3xN] positions with all the relevant points for the device (tip and entry for SEEG depth electrode, 4 corners for ECOG grid). * '''Color''': [1x3] color vector. * '''ContactNumber''': Number of SEEG/ECOG contacts on this device. * '''ContactSpacing''': Distance between the centers of two contacts in the electrode (meters). * '''ContactDiameter''': Diameter of the yellow cylinders representing the contacts (meters). * '''ContactLength''': Length of the yellow cylinder representing the contacts (meters). * '''ElecDiameter''': Diameter of the cylinder representing the SEEG depth electrode (meters). * '''ElecLength''': Length of the cylinder representing the SEEG depth electrode (meters). * '''Visible''': 0 or 1, indicates in the electrode should be rendered in the figures. == Scripting == The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_ecog.m|brainstorm3/toolbox/script/tutorial_ecog.m]] <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:500px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_ecog.m")>><<HTML(</div >)>> <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/Ecog)>> |
== Additional documentation == ==== Related tutorials ==== * [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG/Epilepsy tutorial]] * [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity|sEEG epileptogenicity maps]] * [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Video-EEG|EEG-Video synchronization]] '''Forum''' * Automated screen captures of electrodes: https://neuroimage.usc.edu/forums/t/34628/2 * Set colors for each contact: https://neuroimage.usc.edu/forums/t/35057 |
ECoG/sEEG tutorial
Authors: Francois Tadel, Marcel Heers.
This tutorial introduces some concepts that are specific to the management of ECoG/sEEG recordings in the Brainstorm environment. It is based on a clinical case from the Epilepsy Center at the University Hospital of Freiburg, Germany.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
NOT FOR CLINICAL USE:
The performance characteristics of the methods and software implentation presented in this tutorial have not been certified as medical devices and should be used for research purposes only.
Contents
Dataset description
License
This tutorial dataset (EEG and MRI data) remains proprietary of the Epilepsy Centre, University Hospital Freiburg, Germany. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the Epilepsy Centre in Freiburg. For questions please contact A. Schulze-Bonhage, MD, PhD: andreas.schulze-bonhage@uniklinik-freiburg.de
Acknowledgements
We would like to thank the patient for providing his dataset for this tutorial, the clinical team of the Epilepsy Center Freiburg for the aquisition of the dataset and Verena Schulte for her help in preparing the dataset for the tutorial.
Clinical description
The patient is a 38yr male who suffered from drug resistant focal epilepsy since his 13th year of age with two different types of focal aware seizures that rarely evolved at nighttime to bilateral tonic-clonic seizures. Seizure type I consisted of daily aware focal seizures with myoclonic or clonic jerks of the right leg that occasionally led to falls. Type II consisted of formication paraesthesias that propagated from the right calf to the hip region. It could be provoked by stress or hot weather and occurred 2-3 times a week. In the presurgical 3T epilepsy MRI no structural abnormality was detected.
Implantation scheme
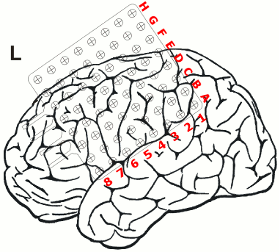
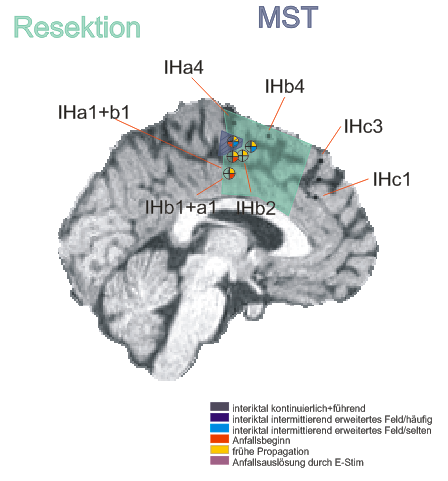
After non-invasive telemetry intracranial recordings were performed. It was decided to implant a 64 channels Ad-Tech subdural grid with 8x8 contacts (stainless steel, 10mm between centers of contacts, 2.3mm diameter exposure) over the left fronto-centro-parietal convexity covering the central region. The most inferior lateral row of contacts of the grid electrode is named A and the row of contacts next to the midline is named H. The contacts of the grid have numbers between 1-8 with the most posterior contacts of each row starting with number 1.
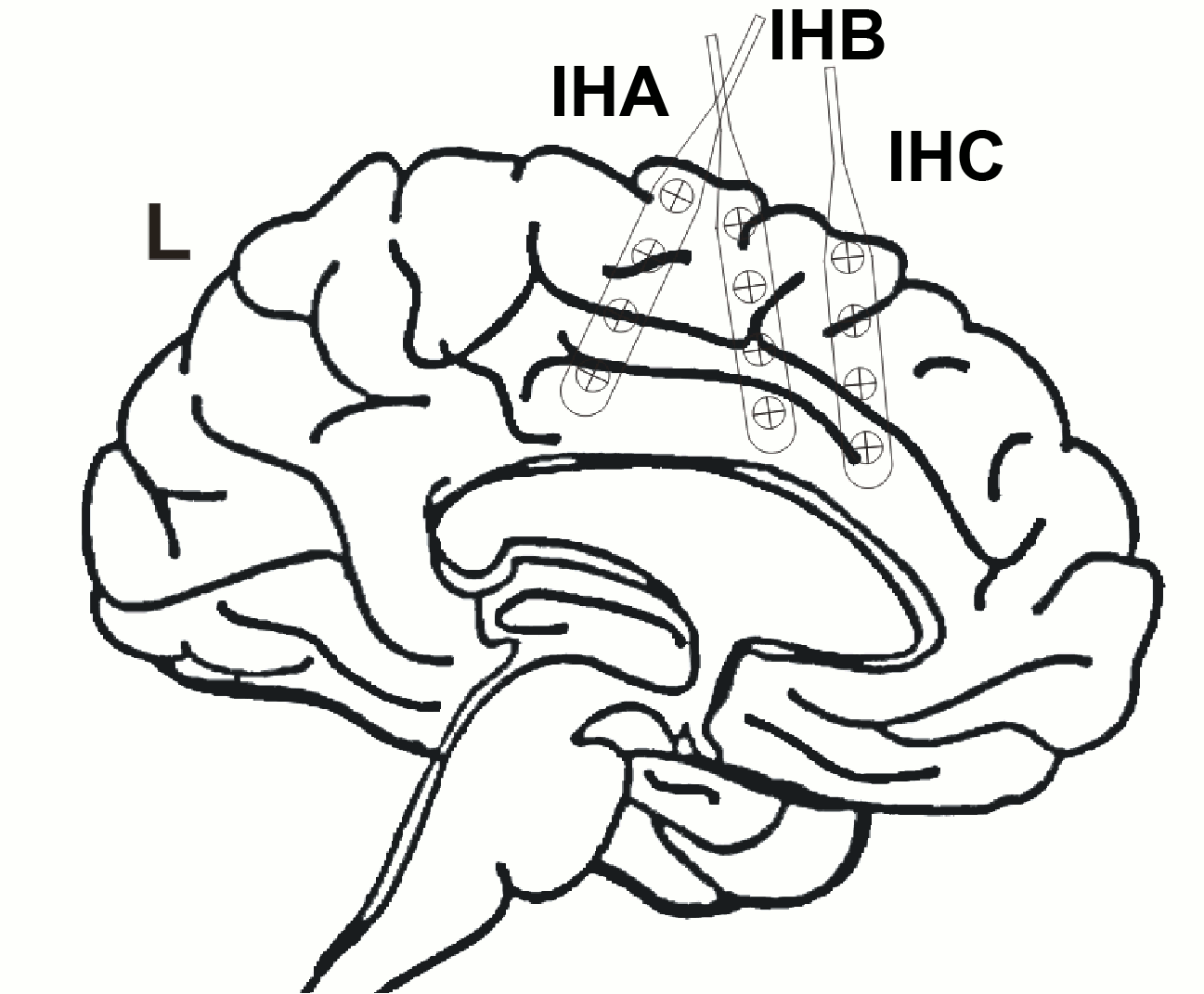
Additionally, 3 interhemispheric Ad-Tech strip electrodes with 4 contacts each named from posterior to anterior with IHA being the most posterior followed by IHB and IHC being the most anterior strip electrode. The numbering convention is that the deepest contact is named contact "1": and the most superficial contact of each electrode has the highest number.
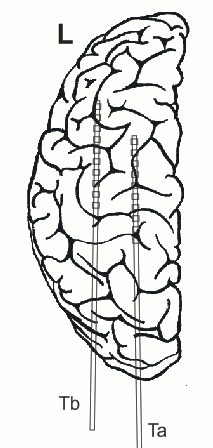
In addition, 2 Ad-Tech sEEG electrodes were implanted in the right parietal lobe facing to the insula (TA & TB, distance between centers of contacts: 10mm, diameter: 0.86mm). Both sEEG electrodes have 10 electrode contacts each. The contacts are counted from anterior to posterior starting with contact number 1 at the tip of each sEEG electrode.
The figure illustrating the electrode positions was created as reported by Kovalev et al. 2005.
Recordings
The EEG data distributed here was recorded at 1024Hz, using a Neurofile NT digital video-EEG system with 128 channels and a 16-bit A/D converter. The signal was filtered in the recording system with a high-pass filter with a time constant of 1 second (cut-off frequency ~ 0.16Hz) and a low-pass filter with a cut-off frequency of 344 Hz.
Clinical evaluation
Interictal findings:
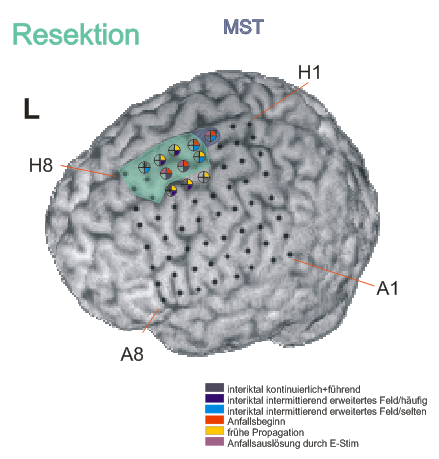
Repetitive sharp waves and interictal fast epileptic activity in the gamma frequency band (FEA; for advanced spectral analysis of FEA see also (Heers et al., 2018)) was almost continuously recorded in contacts G5-6 extending towards H5-6 or F5-6 and less frequently expanding to H3-4 & 7, IHA1-3, IHB1-3
Ictal findings:
- Seizure onset pattern: Repetitive gamma bursts interrupted by sharp slow waves in G5-6, H3-4 and IHA1-3
- Semiology: Myoclonic and clonic movements of the right leg intermingled with not clearly localizing habitual formication paraesthesias of the right leg
Electrical stimulation:
- Stimulation of contacts G5: habitual myoclonic seizures
- Stimulation of contact G6: habitual paraesthesias of the right leg
- Detailed function mapping was performed, which is not reported in detail here.
Resection, histopathology and postsurgical outcome:
- Resection of the posterior part of the superior frontal gyrus anterior of the precentral gyrus including the cingulate gyrus was performed. Because there was overlap between the primary motor cortex (confirmed by functional mapping) at contact H3 position H3, which was also involved in the seizure onset zone this brain area was spared and only multiple subpial transections were added here.
- Histological examination revealed focal cortical dysplasia type Ib (Palmini et al., 2004). After surgery the patient developed a severe SMA syndrome with diminished activity, problems in movement initiation and speech production from which he recovered completely after rehabilitation.
- After surgery the patient remained completely seizure free (Engel IA outcome, follow up 10 years).
References
Heers M, Helias M, Hedrich T, Dümpelmann M, Schulze-Bonhage A, Ball T (2018): Spectral bandwidth of interictal fast epileptic activity characterizes the seizure onset zone. NeuroImage Clin 17.
Kovalev D, Spreer J, Honegger J, Zentner J, Schulze‐Bonhage A, Huppertz HJ (2005): Rapid and fully automated visualization of subdural electrodes in the presurgical evaluation of epilepsy patients. AJNR Am J Neuroradiol 26:1078–1083.
Palmini A, Najm I, Avanzini G, Babb T, Guerrini R, Foldvary-Schaefer N, Jackson G, Luders HO, Prayson R, Spreafico R, Vinters H V (2004): Terminology and classification of the cortical dysplasias. Neurology 62:S2-8.
Files
The dataset we distribute with this tutorial follows the Brain Imaging Data Structure (BIDS) standard for neuroimaging data organization. This specification was first established for MRI and fMRI (Gorgolewski, 2016) and then refined with an extension dedicated to iEEG (Holdgraf, 2019). The files that will be imported in this tutorial are the following:
sample_ecog/
derivatives/: Everything that cannot be considered as raw data
freesurfer/sub-ecog01_ses-preimp/: Result of the FreeSurfer 7.1.1 segmentation for subject ecog01 (T1 pre-implantation)
sub-ecog01/: Raw data for subject ecog01
ses-preimp/: Imaging exams performed before the implantation of the ECoG/sEEG.
anat/sub-ecog01_ses-preimp_T1w.nii.gz: T1-weighted MRI pre-implantation
ses-postimp/: Exams performed with the sEEG/ECoG devices implanted.
anat/sub-ecog01_ses-postimp_T1w.nii.gz: T1-weighted MRI post-implantation
ieeg/..._task-seizure_run-01_ieeg.eeg: 1 hour ECoG+sEEG recordings with 3 seizures (saved using the BrainVision file format, with the header files .vhdr and .vmrk)
ieeg/..._space-IXI549Space_electrodes.tsv: Position of the contacts in MNI space (SPM12 Segment non-linear normalization)
ieeg/..._space-other_electrodes.tsv: Position of the contacts in world coordinates
All the anatomical images have been de-identified with mri_deface from FreeSurfer 6.
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
SPM: The coregistration between pre- and post-implantation MRI volumes requires the SPM12 toolbox. If you are running Brainstorm from a Matlab installation, SPM can be installed or configured as a plugin: menu Plugins > SPM12 > Install. If you are using the stand-alone compiled version of Brainstorm: all the needed SPM scripts have been compiled and included in this executable.
Download the dataset:
Go to the Download page of this website, and get the file: sample_ecog.zip
- Unzip it outside of any of the Brainstorm folders (program folder or database folder)
Brainstorm:
- Start Brainstorm (Matlab scripts or stand-alone version).
Select the menu File > Create new protocol. Name it "TutorialEcog" and select:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
Import the anatomy
The entire dataset, anatomy and recordings, is organized following the BIDS specification. It could be loaded automatically with the menu Import BIDS dataset, as illustrated in the Resting/OMEGA tutorial. However, in this tutorial we will detail the step-by-step instructions to load the anatomy and ECoG recordings manually.
Pre-implantation MRI
The pre-implantation T1 MRI is available as a .nii file (sample_ecog/sub-ecog01/ses-postimp/anat). This volume was processed with FreeSurfer 7.1.1, and the output segmentation is available in the derivatives folder (sample_ecog/derivatives/freesurfer/sub-ecog01_ses-preimp). We don't need to import both to the Brainstorm database: in order to minimize the number of operations, we will import only the FreeSurfer folder.
- Switch to the "anatomy" view of the protocol.
Right-click on the TutorialEcog folder > New subject > ecog01
- Keep the default options you set for the protocol
Right-click on the subject node > Import anatomy folder (auto): More information
Set the file format: FreeSurfer + Volume atlases
Select the folder: sample_ecog/derivatives/freesurfer/sub-ecog01_ses-preimp
At the end, make sure the file "cortex_15000V" is selected (downsampled pial surface), otherwise double-click to select it as the default cortex surface.
Rename the file MRI into T1pre, for better readability in the rest of the tutorial.

When processing your own datasets: Instead of using FreeSurfer, you can decide to run the segmentation of the pre-implantation MRI directly from Brainstorm, using CAT12.
For help with using the volume atlases, refer to the introduction tutorial Explore the anatomy.
Post-implantation MRI
- The pre-implantation MRI will be used as the anatomical reference for this subject. We will now import a second scan done after the sEEG/ECoG implantation, on which we can see the recording contacts. In this dataset, the post-implantation volume is another T1 MRI scan (contacts hyposignal appears in black), but it is more commonly a CT scan (contacts hypersignal appears in white).
Right-click on the subject node > Import MRI:
Select: sample_ecog/sub-ecog01/ses-postimp/anat/sub-ecog01_ses-postimp_T1w.nii.gzDo you want to apply the transformation to the MRI file? YES
This will reorient the MRI in Brainstorm's standard orientation, so you can see the coronal/sagittal/axial views correctly oriented.How to register the new volume? SPM
For more information about these options, see the section Volume coregistration. Note that you need to have the SPM12 toolbox installed on your computer and added to your Matlab path for using this registration method.Reslice the volume? NO
This operation rewrites the volume with the orientation and resolution of the pre-implantation MRI, so that the two volumes can be overlaid in the MRI viewer. In this tutorial, we are going to place the ECoG/sEEG contacts manually on this post-implantation volume. For a better image quality, it is better to do it using the non-resliced volume. We do want the resliced volume, at least to check that pre- and post-implantation MRI volumes are correctly registered, but we can compute the resliced volume separately.Rename the new volume sub-ecog01_ses-postimp_T1w_spm into T1post_spm.

Right-click on the T1post volume > Register with default MRI > Reslice / world coordinates.
Right-click on the resliced T1post volume > Display > Overlay on default MRI
The MRI viewer opens, showing the post-implantation volume as a colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section "Data options" of the Surface tab, adjust its colormap with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well.
Generate skull surfaces
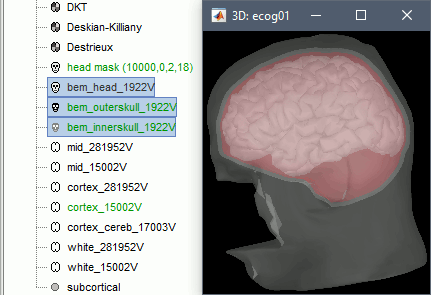
- Skull surfaces are useful for defining the position of ECOG contacts, and displaying them in 3D views. The contacts are projected on the inner skull surfaces, and then they are represented as cylinders oriented tangentially to the skull.
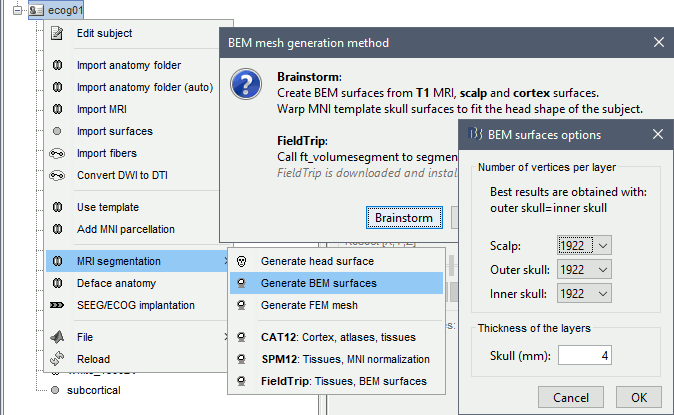
Right-click on the subject folder > Generate BEM surfaces > Brainstorm. Keep the default options.
When the computation is done, double-click on the the original head surface ("head mask") to select it as the default instead of the new BEM version, which is not estimated well because the volume was defaced.
Access the recordings
Link the recordings
- Switch to the "functional data" view (2nd button, on top of the database explorer).
Right-click on the subject folder > Review raw file:
Select the file format: "ECOG: Deltamed/Micromed/..."
Select the recordings: sample_ecog/sub-ecog01/ses-postimp/ieeg/..._ieeg.eeg
The new "Link to raw file" entry lets you access directly the contents of the original file. The menu "Review raw file" does not actually copy any data to the database. More information.
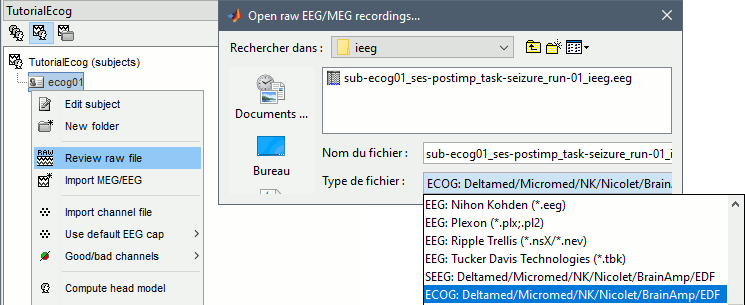
Edit the channel types
At the previous step, we explicitly defined that we were importing ECoG recordings. However, this file contains both sEEG and ECoG recordings, therefore we need to modify manually the type of some channels.
Right-click on the channel file > Edit channel file.
Select all the TA and TB channels, right-click > Set channel type: enter SEEG.
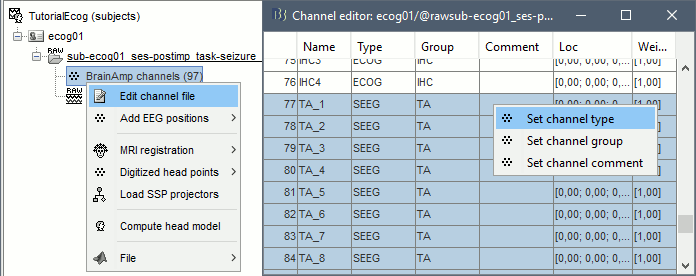
Edit the contacts positions
For 3D mapping and to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the grids and sEEG electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. This tutorial will now explain how to do this manual marking with the MRI Viewer from the post-implantation MRI, which shows clear hyposignal artifacts around the ECoG and sEEG contacts. We prefer using the non-resliced volume because it offers a clearer view of these artifacts.
If you have no interest in this or if you want to skip faster to the analysis of the signals, you can load the 3D positions from the BIDS dataset: read the advanced section Read BIDS electrodes.tsv.
ECoG grid: G
Right-click on the channel file > MRI registration > Edit (MRI Viewer: T1post_spm).
- Adjust the brightness and contrast of the colormap in order to see the contacts artifacts correctly:
- right-click on the figure and move your mouse up/down, or
- left-click on the colormap and move your mouse up/down (brightness) or left/right (contrast), or
right-click on the figure > Colormap:Anatomy (figures below: contrast=100, brightness=100)
For help with all the options iEEG tab, refer to the Epileptogenicity tutorial.
Related tutorial: Edit implantation without recordings.In the iEEG tab, select "G", and select the information we know about this grid:
Type: ECoG
Number of contacts: 8 8 (8x8 grid, for a total of 64 contacts)
Contact spacing: 10 mm
Contact height: 1 mm
(the grid is thinner, but giving it some thickness makes the figures easier to read)Contact diameter: 4 mm
(Ad-Tech contacts have 4mm diameters, with a 2.3mm exposure)- Wire width: Keep the default value, or adjust it later to your taste
- ECoG grids can be defined by pointing successively at their 4 corners. In this example, the contacts of the grid G are labelled sequentially: A1 A2 .. A8 B1 B2 .. B8 .. .. H1 H2 .. H8. You should point at the 4 corners starting by the first two ones, then finish rotating in the same direction: A1, A8, H8, H1.
Navigate in the volume and position the cursor at the center of the artifact corresponding to contact G_A1 (look at the implantation schemes above, it is the more frontal and mesial in the grid), then click on button [#1] in the iEEG tab.
If you can't find this point, you could enter manually the coordinates of the point: click on the button "Set current coordinates" at the bottom of the MRI Viewer window, and copy the MRI coordinates from the screen capture below.
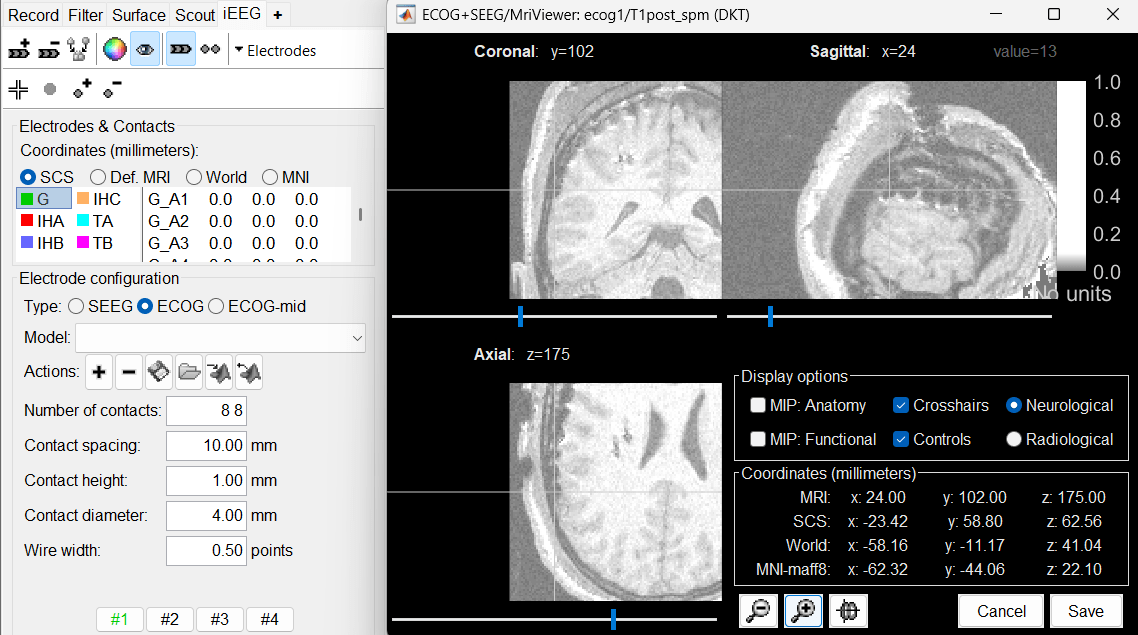
Zoom in/out with the buttons or the associated shortcuts (Ctrl+Scroll or +/-) and explore the volume with (Shift+)x/y/z. Additional display options are available in the popup menu for this figure. All the shortcuts are listed in this tutorial.
Place the cursor at contact G_A8 and click on button [#2].
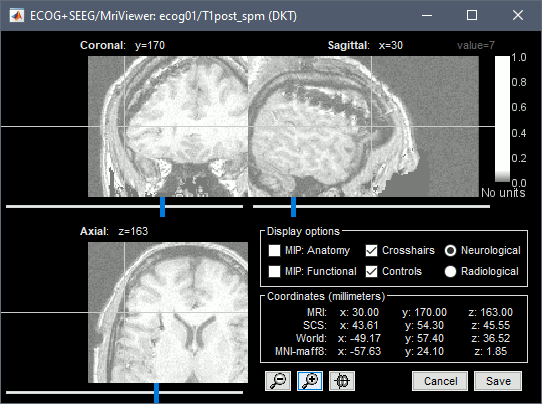
Place the cursor at contact G_H8 and click on button [#3].
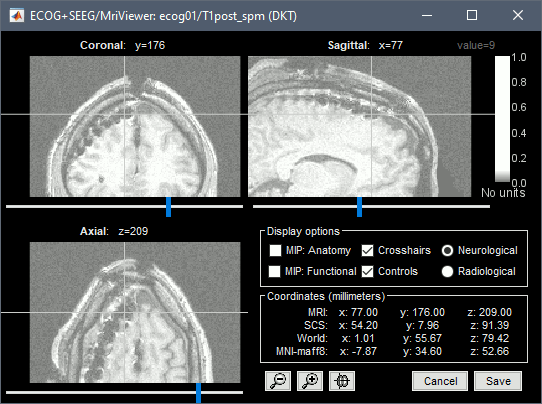
Place the cursor at contact G_H1 and click on button [#4].
After placing the last corner, it should start computing for a while. The positions of the 64 contacts are distributed in the polygon you defined, and then adjusted to the shape of the brain. To get the figure below, with all the contacts projected on all slices, select the checkbox "MIP: Functional" in the MRI Viewer.
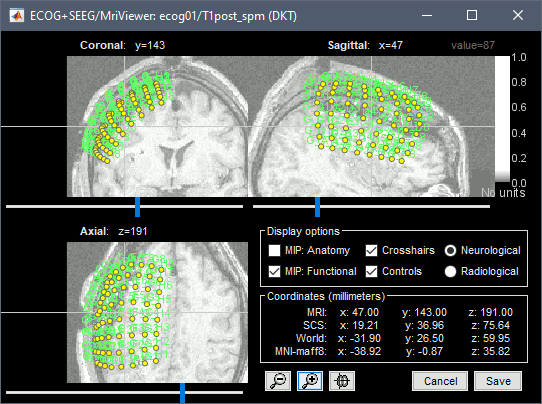
In this window, the contacts can be moved by simple drag-and-drop of the dots representing them. If you move them by mistake, there is unfortunately no undo button. But you can cancel your modifications in two ways: close the figure and discard the modifications, or select "G" in the iEEG tab and then use the menu Contacts > Use default positions, to recompute the initial positions from the 4 four corners of the ECoG grid.
In the same menu Contacts, you can find additional projection options: Project on inner skull and Project on cortex. All these menus work well only when skull surfaces are available for the patient (see the section above that explains how to use the menu Generate BEM surfaces).
From time to time, you can click on the menu Contacts > Save modifications in the iEEG tab. This would prevent you from losing your work if anything unexpected happens.
ECoG strips: IHA, IHB, IHC
IHA
In the iEEG tab, select strip IHA and edit the options:
Type: ECOG-mid (for strips or grids that are not in aligned with the skull surface)
Number of contacts: 4
Contact spacing: 10 mm
Contact height: 1 mm
Contact diameter: 4 mm
- ECoG strips are defined only with two points, the first and the last contact. The intermediate contacts are uniformly distributed between the first and last point. In the convention used here, the first contact is always the deepest one.
If you want to hide the contacts of previously edited devices: select the item in the list at the top of the iEEG tab, then click on the button Show/hide selected electrodes just above the list.
Place the cursor on IHA1 (the deepest contact on the most posterior strip) and click [#1]. To jump directly to the coordinates of this point: click on the button Set current coordinates at the bottom of the MRI viewer, and report the MRI coordinates displayed in the screen capture below.
Place the cursor on IHA4 and click [#2].
Because you have selected the type ECOG-mid, the contacts are not be aligned on the skull or the cortex automatically. The contacts between the first and the last one are by default distributed uniformly on a straight line, which probably doesn't match their actual positions. Move slightly contacts IHA2 and IHA3 so that they appear in the middle of the artifacts.
IHB
Repeat the same steps with IHB: Set it as ECOG-mid, contact spacing=10mm.
Place the cursor on IHB1 (the deepest contact on the middle strip) and click [#1].
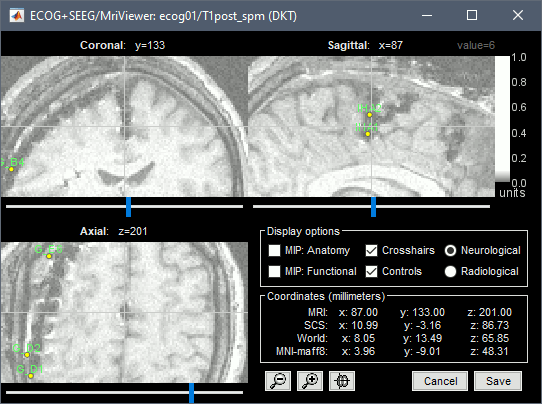
Place the cursor on IHB4 and click [#2].
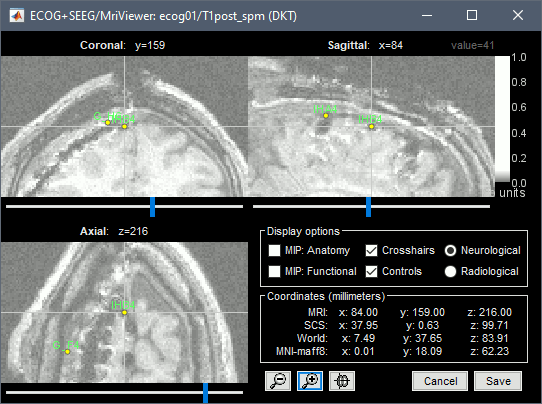
Move the IHB2 and IHB3 contacts on the slices to center them on the artifacts.
IHC
Repeat the same steps with IHC: Set it as ECOG-mid, contact spacing=10mm.
Place the cursor on IHC1 (the deepest contact on the anterior strip) and click [#1].
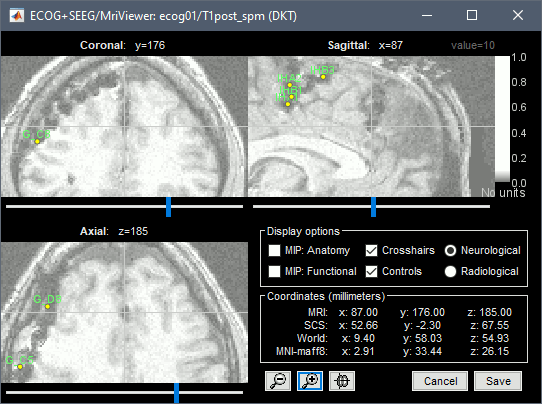
Place the cursor on IHC4 and click [#2].
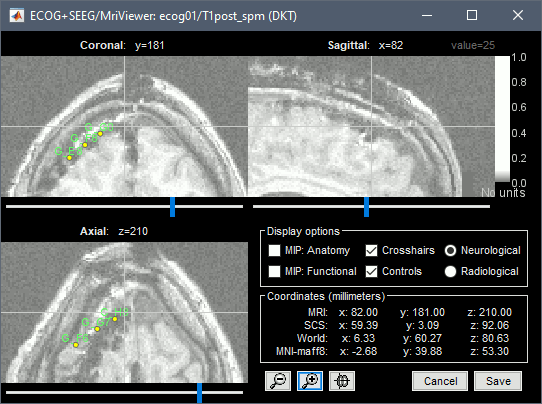
Move the IHC2 and IHC3 contacts on the slices to center them on the artifacts.
sEEG electrodes: TA, TB
TA
In the iEEG tab, select electrode TA and edit the options:
Type: SEEG
Number of contacts: 10
Contact spacing: 10 mm
Contact length: 2.5 mm
Contact diameter: 1 mm
Electrode diameter: 0.9 mm
Electrode length: 120 mm
- The contacts actually have the same diameter as the electrode itself, but we use a slightly higher diameter for display, so that they render well in most 3D and 2D views.
- sEEG electrodes are defined with two points, the tip of the electrode (deepest contact) and a second point anywhere along the axis the electrode. One typical solution is to use the entry point in the skull as this second point, because the screws are always easy to identify. In this example, the electrodes are curved, and the entry point is not aligned with most of the contact, therefore we suggest to pick the last contact on the electrode instead. The intermediate contacts are placed along electrode, separated according to the "contact spacing" option, starting from the tip.
Place the cursor on TA1 (the deepest contact on the more mesial probe) and click [Set tip]. To jump directly to the coordinates of this point: click on the button Set current coordinates at the bottom of the MRI viewer, and report the MRI coordinates displayed in the screen capture below.
Place the cursor on TA10 (the last contact on this electrode) and click [Set skull entry].
This is obviously not the location of the screw in the skull, but as the electrode is bent, this is a better point to approximate the contact locations on a straight line.
The electrode is curved, therefore you need to move most of the intermediate contacts TA2-TA9 to more appropriate positions, at the center of the matching artifact. Unfortunately, Brainstorm does not handle well curved electrodes yet: if you move the contacts, the electrode remains represented as a straight line. This is a display issue only, which has no impact on any computation, but we hope to improve it at some point.
TB
Repeat the same steps with TB. with the same configuration:
Type: SEEG
Number of contacts: 10
Contact spacing: 10 mm
Contact length: 2.5 mm
Contact diameter: 1 mm
Electrode diameter: 0.9 mm
Electrode length: 120 mm
Place the cursor on TB1 (the deepest contact on the more mesial probe) and click [Set tip].
Place the cursor on TB10 (the last contact on this electrode) and click [Set skull entry].
Move the intermediate contacts TB2-TB9 to the centers of the artifacts.
When you are done with marking all the electrodes, click on [Save] in the MRI viewer.
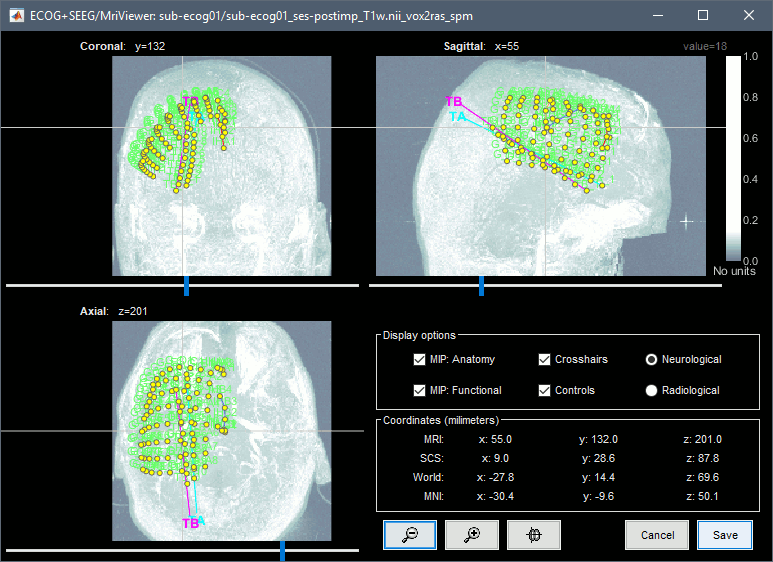
3D display
Right-click on the channel file > Display sensors > Explore all the options available.
For help with the display options in the iEEG tab: Epileptogenicity tutorial.
You can add more anatomy elements to the figure with the button "Add a surface" at the top-right of the Surface tab. For more help: Display the anatomy.
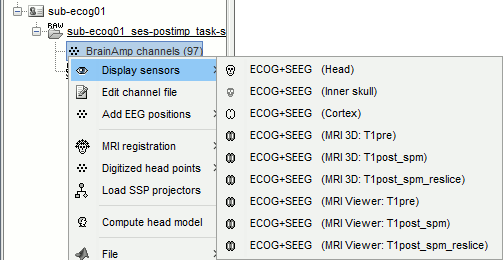
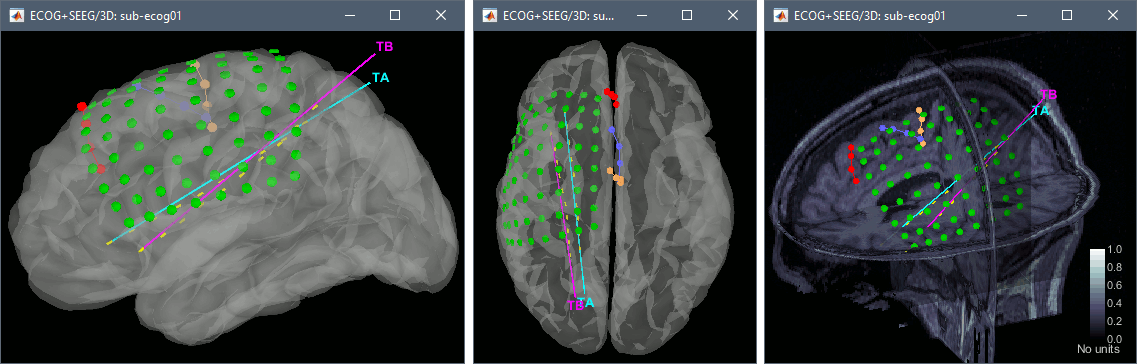
Project on brain surface
You probably noticed that many the contacts of the ECoG grid appear under the surface of the cortex in these 3D figures. This is mostly due to the brain shift between the volumes we used to mark the contacts (post-implantation MRI) and the volume from which the cortex surface was reconstructed (pre-implantation MRI). One solution to compensate for this brain shift and produce more readable figures is to project the contacts on the pre-implantation surfaces.
In the iEEG tab, select the grid G, then use menu Contacts > Project on inner skull.
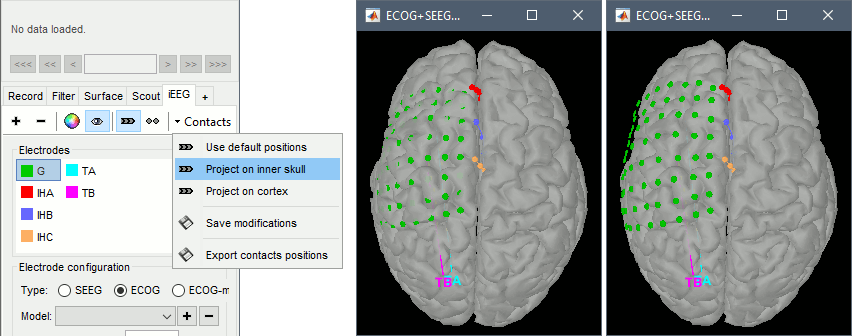
You could also test what you obtain by projecting the contacts on the inner skull. It produces similar results, but with a different method, projecting the contacts possibily a bit further away from the brain. Depending on the quality of the surfaces, the shape of the brain, and the position of the contacts, one solution may produce weird results, try the other one in that case.
Alternative: Read BIDS electrodes.tsv
A BIDS dataset may contain information about the localization of the iEEG contacts in different systems of coordinates. These files are documented in the in BIDS specification. In this dataset, we can find the following files in the folder sample_ecog/sub-ecog01/ses-postimp/ieeg. Files
sub-ecog01_ses-postimp_space-IXI549Space_coordsystem.json: MNI space metadata
sub-ecog01_ses-postimp_space-IXI549Space_electrodes.tsv: XYZ coordinates, MNI space
sub-ecog01_ses-postimp_space-ScanRAS_coordsystem.json: Subject space metadata
sub-ecog01_ses-postimp_space-ScanRAS_electrodes.tsv: XYZ coordinates, subject space
The tag space- indicates how the coordinates should be interpreted (list of possible values):
IXI549Space: Refers to a MNI normalization computed by SPM12 (see Coordinate systems). You should be able to load these locations on any volume for which MNI coordinates are available, but with the approximations due to the volume-based normalization and possible errors due to the use of different types of normalization. In this particular dataset, the MNI coordinates were computed from Brainstorm with the SPM12 Segment non-linear normalization. When possible, always prefer importing coordinates in subject space.
ScanRAS: Refers to the scanner coordinates (named world coordinates in Brainstorm) of a specific anatomical image (MRI or CT). The interpretation of the coordinates in the file electrodes.tsv requires the information available in the adjacent file coordsystem.json. Typically, this pair of files defines coordinates associated to a specific volume in the BIDS dataset.
For the subject space (space-ScanRAS), the contents of the json file is the following:
"iEEGCoordinateSystem": "ScanRAS", "iEEGCoordinateUnits": "mm", "iEEGCoordinateSystemDescription": "Scanner-based RAS coordinates of the post-implantation T1 MRI", "iEEGCoordinateProcessingDescription": "SEEG contacts manually placed with Brainstorm on the post-implanation T1 MRI", "iEEGCoordinateProcessingReference": "https://neuroimage.usc.edu/brainstorm/Tutorials/ECoG", "IntendedFor": "sub-ecog01/ses-postimp/anat/sub-ecog01_ses-postimp_T1w.nii.gz"
The field IntendedFor points at the file sub-ecog01_ses-postimp_T1w.nii.gz. This means that the coordinates in the files sub-ecog01_ses-postimp_space-ScanRAS_electrodes.tsv refer to the post-implantation T1 MRI.
To add the coordinates in subject space (be careful: if you already marked the contacts manually, you would lose all your work - if you want to try this, duplicate the subject first):
Right-click on the channel file > Add EEG positions > Import from file
Select format "EEG: BIDS electrodes.tsv, subject space mm"
(or adjust depending on the tsv file you select and the information defined in the json file)Select file sub-ecog01_ses-postimp_space-ScanRAS_electrodes.tsv
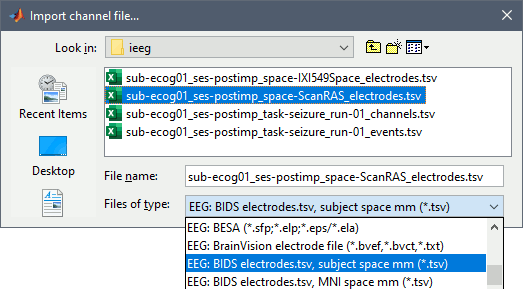
Note that the electrodes.tsv file does not include all the information we defined manually in the previous section (ECOG vs. ECOG-mid, contact diameter...), therefore for optimal display you might need to go through the configuration of each implanted device in the iEEG tab and edit the values as previously.
Display the recordings
ECoG/sEEG time series
Right-click on any "Link to raw file" > ECOG+SEEG > Display time series.
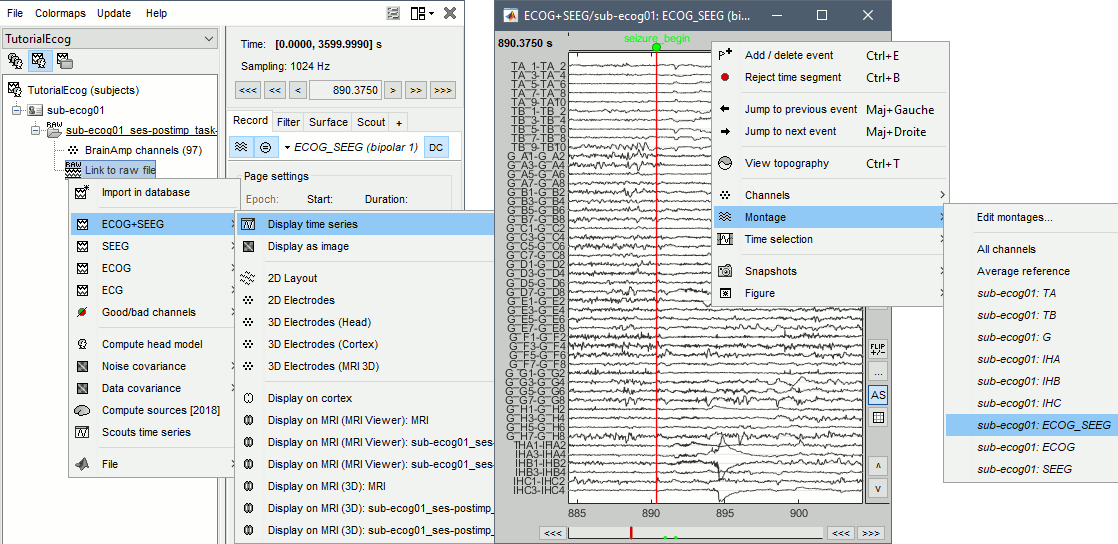
- For instructions on how to use this viewer, please refer to these tutorials:
- Typical display configuration includes:
Set the page duration to 20s or 30s: Record tab > Page settings.
Change the display mode to "columns": Record tab > Toolbar > First button.
Change the montage (bipolar/monopolar): Record tab > Drop-down menu.
- Set visualization filters: Filter tab.
- Disable the automatic scaling when changing page: Button [AS] in the time series figure.
- Adjust the amplitude scale: Right-click+move up/down, or buttons [^] and [v].
Set the resolution precisely: Right-click on the figure > Figure > Set axes resolution.
- Montage options include, for each group separately or all the channels together:
Bipolar 1: Difference of pairs of adjacent sensors (sparse), eg. A1-A2, A3-A4, A5-A6
Bipolar 2: Difference of pairs of adjacent sensors (exhaustive), eg. A1-A2, A2-A3, A3-A4
Local average reference: Average reference computed independently for each group
2D/3D topography
With the menus 2DLayout, 2D Electrodes and 3D Electrodes you can visualize the ECoG/sEEG values at the current time point. Move the time cursor and it will update the values represented on the contacts.
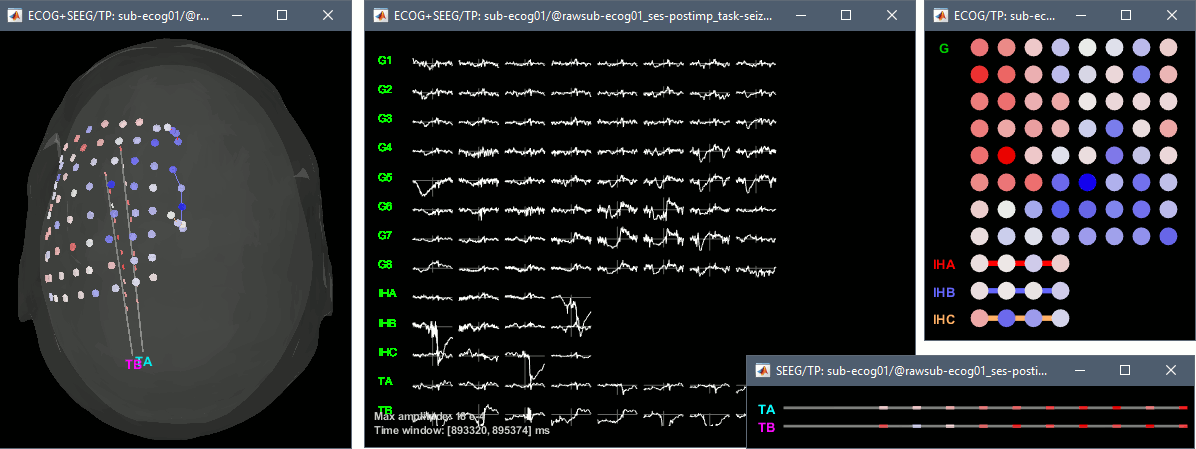
Interpolate on the anatomy
From the same ECOG+SEEG display menu, you can also project the recordings on the MRI or the cortex. You can edit the colormap for the displayed values like any other colormap.
MRI volume: All the voxels in the neighborhood of a contact are attributed the value associated with this contact. In the figure below, both "MIP: functional" and "MIP: anatomy" options are selected (maximum intensity projection along each axis = "glass brain" view).
Cortex surface: The values of the vertices are interpolated from the nearest SEEG contacts (the magnitude decreases with the distance to the contact).
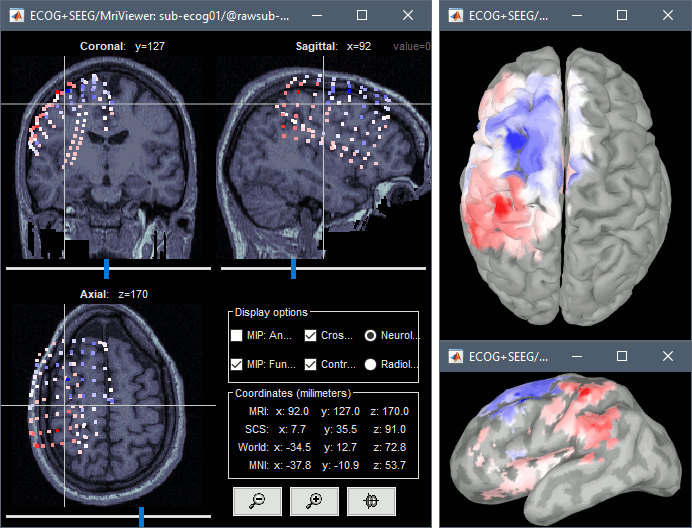
Power spectrum
We recommend you always start your data analysis with a spectral evalution of the recordings, it may help you identify bad channels. This is described in tutorials Power spectrum and EEG and epilepsy.
- In Process1, select the continuous file.
Run process Frequency > Power spectrum density (Welch): All file, Length=10s, ECOG,SEEG.
Double-click on the PSD files to display them, change to a log-scale for the X axis, zoom in. None of the channels seem to have noise levels that are higher or lower than the others, all the channels are possibly good.
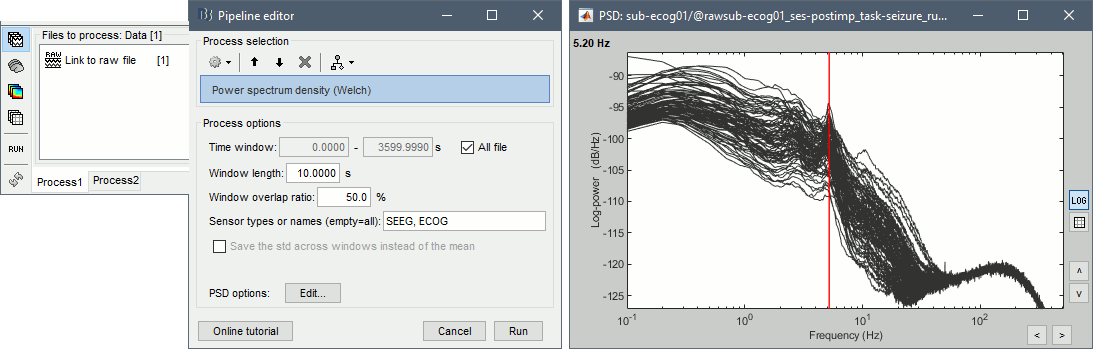
- If the spectrum plots are too noisy and difficult to read, you can recompute them with lower frequency resolution. Decrease the duration of the estimator window, possibly down to 1s depending on the sampling frequency. This will estimate the PSD by averaging the power of more time windows with less data bins, the result will be smoother.
Import epochs of interest
At this point of the analysis, we are still looking at the original files, no data was copied to the database. The montages are saved in the Brainstorm preferences, the bad channels and new events are saved in the links of the database but not reported to the original files. If you delete your protocol at this point, you would only lose the event marking and bad channel selection.
We are now going to import a segment a recordings around each of the three seizures marked in this file (10s before and 20s after the onset). This will make real copies of the data in the database, so we can run additional processes on them.
- Double-click on the on the "Link to raw file" to open the time series figure (ECOG+SEEG).
In the Record tab: double-click on the event category seizure_begin and rename it seizure.
- Close the file viewer to save the modification to the events.
Right-click on Link to raw file > Import in database:
- Time window: All
- Split in time blocks: Disabled
Use events: Enabled, select seizure
Epoch time: [-10000, +20000] ms (imports -10s to 20s around the event "seizure")
- Remove DC offset: Disabled
- Resample: Disabled
Create a separate folder for each event type: Disabled
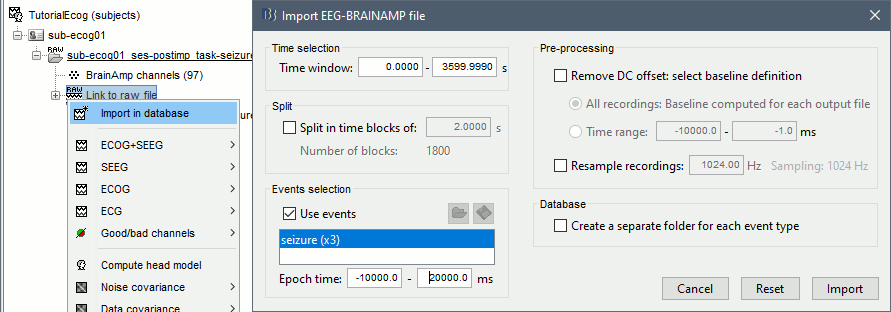
- At the end, you should have three new folders SZ1/SZ2/SZ3, the same name as the original .TRC files, but without the tag "raw" on top. These new folders contain copies of the SEEG recordings, if you delete these folders from the database explorer, you lose the recordings they contain.
The imported epochs are saved with a new timing: for the seizure onsets, the reference time t=0s is now the event "Onset", which has been removed from the list. You can still see the other marker Seizure, which should be slightly before 0s. For the baselines, the reference t=0 is the beginning of the segment.
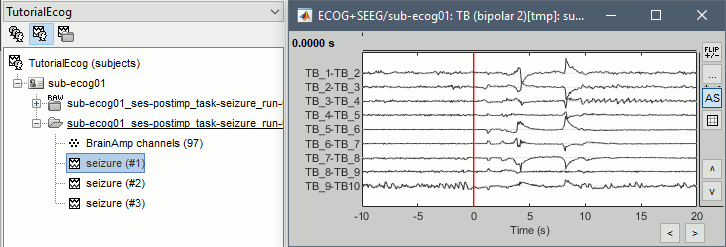
Anatomical labelling
As illustrated in the introduction tutorial Explore the anatomy, all the anatomical atlases available in the subject (e.g. DKT, Desikan-Killiany, Destrieux) are available in the MRI Viewer. Select the current atlas from the figure popup menu, and the anatomical label under the cursor is displayed at the top-right corner. You can overlay all the labels on the MRI with the menu Show atlas.
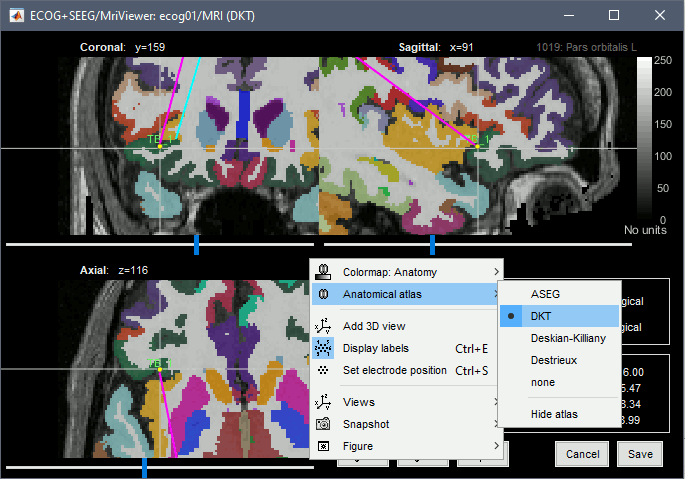
We can use these anatomical parcellations, and also the surface parcellations of the cortex available as scouts, in order to label automatically all the ECOG/SEEG contacts.
If accurate MNI coordinates are expected, it is recommended to use a non-linear MNI normalization instead of the initial linear registration computed at the beginning of this tutorial, as illustrated in the SEEG epileptogenicity tutorial. In the anatomy view, right-click on the pre-implantation MRI (first file in the subject anatomy) > MNI normalization > Segment. This requires SPM12.
Switch to the functional view. Right-click on the channel file > iEEG atlas labels > Select all the available options: XYZ coordinates in various coordinate systems, volume parcellations, surface parcellations.
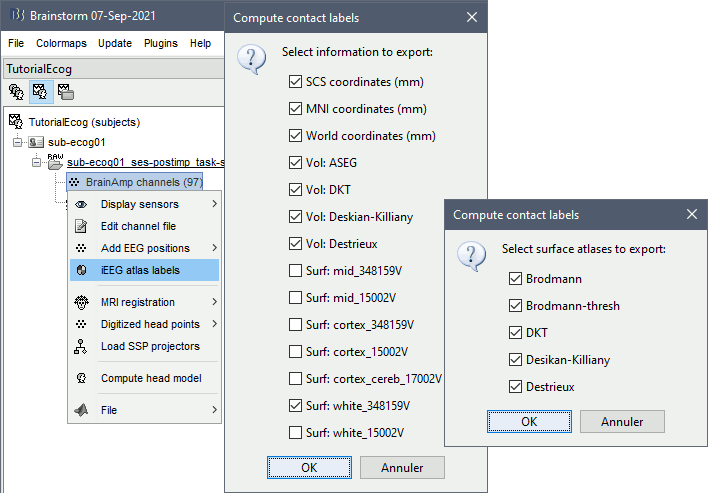
When using surface atlases, select preferencially the high-resolution surfaces (~300000V): a higher density of labelled points on the surface will increase the chances to label correctly the contacts. Selecting the type of surface ("white", "mid" or "cortex") depends on multiple factors, and we don't have enough experience yet to give clear recommendations. To help you decide, here is what these labels mean:
White: Represents the interface between the white matter and the gray matter
Cortex: Inflated from the white surface, represents the interface between the gray matter and the CSF.
Mid: In the FreeSurfer case, this surface is computed by Brainstorm as the layer half-way between the white and the cortex surfaces. If this is available, this might be your best guess. Try it yourself and please share your results and comments on the user forum.
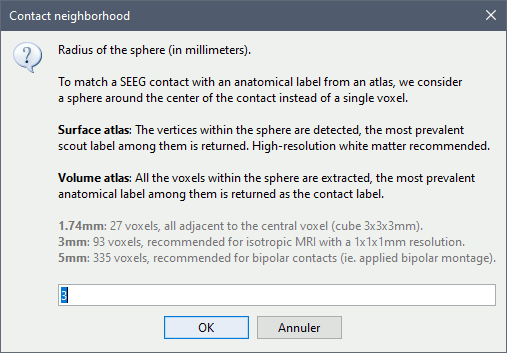
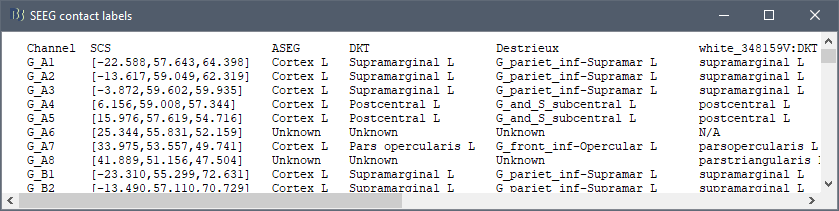
More options and details in the tutorial SEEG epileptogenicity maps.
Additional documentation
Related tutorials
Forum
Automated screen captures of electrodes: https://neuroimage.usc.edu/forums/t/34628/2
Set colors for each contact: https://neuroimage.usc.edu/forums/t/35057
