|
Size: 10896
Comment:
|
Size: 11271
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from brainsuiteBDP ## page was renamed from brainsuite |
'''[TUTORIAL UNDER CONSTRUCTION: NOT READY FOR PUBLIC USE]''' ---- |
| Line 6: | Line 8: |
| '''[TUTORIAL UNDER WRITING: NOT READY FOR PUBLIC USE]''' | In this tutorial, we describe the estimation of realistic conductivity tensors of living brain tissues using the [[http://brainsuite.org/|BrainSuite software]]. These results are used in FEM forward modeling, as described in the tutorials: [[https://neuroimage.usc.edu/brainstorm/Tutorials/Duneuro#DUNEuro_options:_Advanced|FEM with DUNEuro]] and [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMedianNerve#FEM_tensors|FEM median nerve example]]. |
| Line 8: | Line 10: |
| In this tutorial, we describe the process of the estimation of the realistic conductivity tensors for the brain tissues using the [[http://brainsuite.org/|BrainSuite software]]. The main purpose is to generate the conductivity tensors for the FEM computation as introduced on [[https://neuroimage.usc.edu/brainstorm/Tutorials/Duneuro|this page]]. | The realistic tensors are estimated from the Diffusion-Weighted Images (DWI): Brainstorm calls the BrainSuite software to compute the diffusion tensors on each brain MRI voxel (DTI), then Effective Medium Approach (EMA) is applied to estimate the conductivity tensors for each element of a tetrahedral FEM mesh. This is particularly interesting for the modeling the anisotropy of the white matter. |
| Line 10: | Line 12: |
| The realistic tensors are estimated from the Diffusion-Weighted Images (DWI). For this purpose, Brainstorm calls internally the BrainSuite software to compute the diffusion tensors on each brain MRI voxel. Afterward, the Effective Medium Approach (EMA) is applied to convert the diffusion tensors to the conductivity tensors. The following section shows the users how to do it from the Brainstorm graphical interface. Further documentation about previous usage of Brainsuite within Brainstorm can be found in this [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegBrainSuite|page]]. |
BrainSuite is also used for other purposes in Brainstorm, particularly the T1 MRI segmentation, as documented in this tutorial: [[Tutorials/SegBrainSuite|MRI segmentation: BrainSuite]]. |
| Line 18: | Line 16: |
| == Requirement == * You have already followed all the introduction tutorials * You have a working copy of Brainstorm installed on your computer * For the DWI data, only the NIfTi files are supported |
== Download and installation == ==== Requirements ==== * You have already followed all the introduction tutorials. * You have a working copy of Brainstorm installed on your computer. * For the DWI data, only the NIfTI files (.nii) are supported. |
| Line 23: | Line 22: |
| == Brainsuite Installation == 1. Download the latest version of BrainSuite from http://www.brainsuite.org/download. |
==== Install Brainsuite ==== 1. Download the latest version of BrainSuite from http://forums.brainsuite.org/download/. |
| Line 28: | Line 27: |
| 1. Note that you will be using BrainSuite Diffusion Pipeline(BDP), so you need to install a compatible [[http://www.mathworks.com/products/compiler/mcr|MATLAB Compiler Runtime]](last version). | 1. You will be using BrainSuite Diffusion Pipeline (BDP), so you need to install a compatible [[https://www.mathworks.com/products/compiler/matlab-runtime.html|MATLAB Runtime]] (2019b for BrainSuite 21a). |
| Line 30: | Line 29: |
| 1. Start BrainSuite to check if the installation (It's not required to open BrainSuite to run this tutorial). | 1. In Brainstorm, menu File > Edit preferences > Enter the BrainSuite installation folder:<<BR>><<BR>> {{attachment:brainsuiteInstall.gif}} |
| Line 32: | Line 31: |
| 1. The BrainSuite installation folder should be informed in the Brainstorm preferences | ==== Download the dataset ==== * Download the files: [[http://brainsuite.org/WebTutorialData/BrainSuiteTutorialSVReg_Sept16.zip|MRI T1w]] and [[http://brainsuite.org/WebTutorialData/DWI_Feb15.zip|MRI DWI]] (from the [[http://brainsuite.org/tutorials/dtiexercise/|BrainSuite diffusion tutorial]]). |
| Line 34: | Line 34: |
| {{attachment:brainsuiteInstalation.JPG||height="250",width="650"}} | * Unzip it outside of any of the Brainstorm folders (program folder or database folder). * Start Brainstorm (Matlab scripts or stand-alone version) * Select the menu File > Create new protocol. Name it "'''TutorialTensors'''" and select: * No, use individual anatomy * No, use one channel file per condition |
| Line 36: | Line 40: |
| ---- == Dataset == In this tutorial, we use the Brainsuite dataset example available on the BrainSuite [[http://brainsuite.org/tutorials/|tutorial webpage]]. User can also download directly these data from these links: [[http://brainsuite.org/WebTutorialData/BrainSuiteTutorialSVReg_Sept16.zip|MRI T1w]] and [[http://brainsuite.org/WebTutorialData/DWI_Feb15.zip|MRI DWI]] |
== Import the anatomy == === T1 MRI === * Switch to the "anatomical data" view, the left button in the toolbar above the database explorer. * Right-click on the TutorialFem folder > New subject > '''Subject01''' * Keep the default options you set for the protocol. |
| Line 40: | Line 46: |
| The first file contains the T1 MRI data, with the name '2523412.nii.gz'. as in this figure, | * Right-click on the subject node > '''Import MRI''': * Set the file format: '''All MRI files (subject space)''' |
| Line 42: | Line 49: |
| {{attachment:vierMriFolder.JPG||height="150",width="650"}} | * Select the T1 file: BrainSuiteTutorialSVReg/'''2523412.nii.gz''' |
| Line 44: | Line 51: |
| The second file is the DWI and should contain at least three files | * Click on the link "'''Click here to compute MNI normalization'''": option "'''maff8'''". This estimates an affine transformation to the [[https://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|MNI space]] and sets default positions for the anatomical fiducials. The NAS/LPA/RPA fiducials are needed for defining the Brainstorm [[CoordinateSystems|subject coordinate system]], in which the surfaces and FEM meshes are stored. <<BR>><<BR>> {{attachment:importT1.gif}} |
| Line 46: | Line 53: |
| {{attachment:vierDWIFolder.JPG||height="180",width="650"}} | === Diffusion imaging === This computes the This requires BrainSuite to be installed on your computer, with the bdp program available in the system path. |
| Line 48: | Line 56: |
| Where the *.bval is a text file that contains the value of the gradient, and the *.bvec is also a text file that contains the orientation of the gradient. The nii.gz file is the NifTi file of the DWI where the images are stored. | * Right-click on Subject01''' '''> '''Convert DWI to DTI''' |
| Line 50: | Line 58: |
| == Realistic condctivity tensors == === Load the T1 MRI data to brainstorm === First, you need to create a new subject in your protocol, let call it the 'BrainSuiteSubject'. Then import the T1 MRI of the subject and set the fiducials points as explained in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|previous tutorial]]. |
* Select the DWI file: DWI/'''2523412.dwi.nii.gz''' |
| Line 54: | Line 60: |
| The T1 MRI is required since all the BDP uses the T1 space for its computation. Furthermore, this is required since it will be used to align the tensors to the FEM mesh later. | * The associated text files '''*.bvec''' (orientation of the gradient) and '''*.bval''' (value of the gradient) must be in the same folder, with the same file name. Theses files are created from for the DWI acquisition. If you don't have them, ask the person who programmed your DWI sequence and get the files that are specific to your use case. |
| Line 56: | Line 62: |
| {{attachment:mri.JPG||height="100",width="250"}} | * The process can take up to 30min. At the end, a new file '''DTI-EIG''' appears in the database (DTI=diffusion tensors images, EIG=eigenvalue). This file contains 12 volumes, ie. 12 values for each voxel. From 1 to 9: components of the three eigenvectors; from 10 to 12: the values of their norm to the eigenvalue. <<BR>><<BR>> {{attachment:importDTI.gif}} |
| Line 58: | Line 64: |
| === Diffusion tensor generation (DTI) from DWI === In this step, Brainstorm calls the Brainsuite internally, and the diffusion tensors are computed. |
== FEM mesh == The FEM approach requires a segmentation of the head volume in different tissues, represented as hexahedral or tetrahedral 3D meshes. The methods available within Brainstorm are listed in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh|FEM mesh generation]]. |
| Line 61: | Line 67: |
| Right-click on the subject and then select the item "Convert DWI to DTI". | Here we illustrate only the use of '''Brain2mesh''': this is not the most accurate solution for MRI segmentation but it is probably the fastest solution to obtain a tetrahedral mesh of the head with 5 tissues (gray matter, white matter, CSF, skull, skin). For more accurate results, we recommend using '''SimNIBS with T1+T2''', as illustrated in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMedianNerve#FEM_mesh_with_SimNIBS|FEM median nerve example]]. |
| Line 63: | Line 69: |
| {{attachment:dwi2dti.jpg||height="300",width="550"}} Then follow the popup windows by selecting the DWI, you may need to extract the zip file before. {{attachment:importDWI.JPG||height="280",width="650"}} If the bval, and bval files are in the same folder, Brainstorm will detect them automatically, otherwise, the user will be asked to browse the files one by one (as it's the case in this tutorial). Brainstorm calls internally the BrainSuite process, and compute the diffusion tensors. At the end of this process, a new node will appear in the Brainstorm database with the name 'DTI-EIT'. This name refers to, DTI: diffusion tensors images, and EIG for eigenvalue, since the eigenvalues and eigenvectors are computed at voxel and stored in Brainstorm database. {{attachment:mriAndDTI-EIG.JPG||height="100",width="230"}} If you check the structure of the file DTI-EIG, by right click -> File and then 'Display file contents', the following figure is displayed. {{attachment:EIG-hardDisc.JPG||height="200",width="450"}} The size of the matrix is 128x256x256x12, where the first 3 values are the same as the size of the T1 MRI and 12 corresponds to the 3 eigenvectors components (9) and eigenvalues (3) |
* Right-click on the T1 MRI > MRI segmentation > '''Generate FEM mesh''' > '''Brain2mesh'''.<<BR>><<BR>> {{attachment:femMesh.gif}} * After less than 15 minutes, you will obtain a new FEM mesh in the database.<<BR>><<BR>>{{attachment:femMesh2.gif}} |
| Line 86: | Line 75: |
| ==== FEM mesh head model ==== This step requires the FEM mesh of the head model. You can generate the FEM head model from the MRI data as explained on [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh|this page]]. |
www.pnas.org/content/98/20/11697 |
| Line 89: | Line 77: |
| For the following, we used the SimNibs FEM mesh generation. The following figure shows the FEM mesh obtained with the SimNibs method using the T1 MRI. | |
| Line 91: | Line 78: |
| {{attachment:Mri&femMeshView.JPG|Mri&femMeshView.JPG|height="300",width="260"}} {{attachment:femMeshView.JPG||height="300",width="280"}} Note that this mesh is obtained only from the T1, the use of the T2 is highly recommended if it's available, as recommended in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh|FEM mesh tutorial]]. |
|
| Line 100: | Line 84: |
| {{attachment:menuGenerateFemTensors.png||height="280",width="250"}} | {{attachment:menuGenerateFemTensors.png||width="250",height="280"}} |
| Line 104: | Line 88: |
| {{attachment:FEMConductivitiesIsoPanel.JPG||height="220",width="250"}} | {{attachment:FEMConductivitiesIsoPanel.JPG||width="250",height="220"}} |
| Line 112: | Line 96: |
| {{attachment:FEMConductivitiesAnisoPanel.JPG||height="300",width="250"}} | {{attachment:FEMConductivitiesAnisoPanel.JPG||width="250",height="300"}} |
| Line 129: | Line 113: |
| {{attachment:menuDisplayTensors.jpg||height="300",width="250"}} | {{attachment:menuDisplayTensors.jpg||width="250",height="300"}} |
| Line 133: | Line 117: |
| {{attachment:meshViewTensorsLines.JPG||height="300",width="350"}} {{attachment:meshViewTensorsTensorsTops.JPG||height="300",width="350"}} | {{attachment:meshViewTensorsLines.JPG||width="350",height="300"}} {{attachment:meshViewTensorsTensorsTops.JPG||width="350",height="300"}} |
| Line 141: | Line 125: |
| {{attachment:meshViewTensorsLinesWM2.JPG||height="300",width="350"}} {{attachment:tensorsOnMri.JPG||height="300",width="300"}} | {{attachment:meshViewTensorsLinesWM2.JPG||width="350",height="300"}} {{attachment:tensorsOnMri.JPG||width="300",height="300"}} |
| Line 144: | Line 128: |
| In the case where the user wants to use generate isotropic tensors, then the DTI is not required. In the case where more than one FEM head model is in the database, the highlighted one in the green color will be used. | In the case where the user wants to use generate isotropic tensors, then the DTI is not required. For that case, keep all the options to 'isotropic', the recommended display is the 'Ellipsoids', and the final shape will be a sphere (isotropic direction). In the case where more than one FEM head model is in the database, the highlighted one in the green color will be used. |
| Line 148: | Line 132: |
| == Artificial/simulated conductivity tensors == | == Simulated conductivity tensor == |
| Line 151: | Line 135: |
| Users can reach this option by following this tutorial and select the third method in this panel. {{attachment:artificialTensors.JPG||height="450",width="280"}} | Users can reach this option by following this tutorial and select the third method in this panel. {{attachment:artificialTensors.JPG||width="280",height="450"}} |
| Line 157: | Line 141: |
| {{attachment:skullAniso.JPG||height="250",width="300"}} {{attachment:skullAniso2.JPG||height="250",width="300"}} | "eigenvalues parallel (longitudinal) and perpendicular (transverse) to the fiber directions" for 1:10 anisotropy (transverse:longitudinal) {{attachment:skullAniso.JPG||width="300",height="250"}} {{attachment:skullAniso2.JPG||width="300",height="250"}} == Troubleshooting == To be completed soon and linked to BrainSuite website |
| Line 160: | Line 149: |
| To be completed soon |
|
| Line 164: | Line 151: |
| Check the error in the simnibs mesh in X direction and overlay on mri Check the error with the brain2mesh Correct the ratio from integer to float add an interactive way yo change the size of the tensor correct the name of the simulated method, correct the EMC and remove the VC and change the coefficcient | Check the error in the simnibs mesh in X direction and overlay on mri check the error with the brain2mesh Correct the ratio from integer to float check the meaning of transversal/longitidunal in the code add an interactive way yo change the size of the tensor.. important correct the name of the simulated method, correct the EMC and remove the VC and change the coefficcient |
[TUTORIAL UNDER CONSTRUCTION: NOT READY FOR PUBLIC USE]
FEM tensors estimation with BrainSuite
Authors: Takfarinas Medani, Francois Tadel, Anand Joshi and Richard Leahy
In this tutorial, we describe the estimation of realistic conductivity tensors of living brain tissues using the BrainSuite software. These results are used in FEM forward modeling, as described in the tutorials: FEM with DUNEuro and FEM median nerve example.
The realistic tensors are estimated from the Diffusion-Weighted Images (DWI): Brainstorm calls the BrainSuite software to compute the diffusion tensors on each brain MRI voxel (DTI), then Effective Medium Approach (EMA) is applied to estimate the conductivity tensors for each element of a tetrahedral FEM mesh. This is particularly interesting for the modeling the anisotropy of the white matter.
BrainSuite is also used for other purposes in Brainstorm, particularly the T1 MRI segmentation, as documented in this tutorial: MRI segmentation: BrainSuite.
Contents
Download and installation
Requirements
- You have already followed all the introduction tutorials.
- You have a working copy of Brainstorm installed on your computer.
- For the DWI data, only the NIfTI files (.nii) are supported.
Install Brainsuite
Download the latest version of BrainSuite from http://forums.brainsuite.org/download/.
Install it on your computer by following the instructions in BrainSuite's quick start installation guide.
You will be using BrainSuite Diffusion Pipeline (BDP), so you need to install a compatible MATLAB Runtime (2019b for BrainSuite 21a).
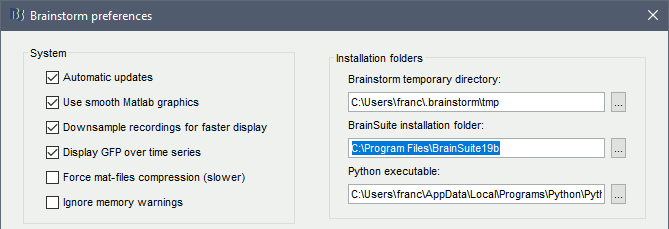
In Brainstorm, menu File > Edit preferences > Enter the BrainSuite installation folder:
Download the dataset
Download the files: MRI T1w and MRI DWI (from the BrainSuite diffusion tutorial).
- Unzip it outside of any of the Brainstorm folders (program folder or database folder).
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialTensors" and select:
- No, use individual anatomy
- No, use one channel file per condition
Import the anatomy
T1 MRI
- Switch to the "anatomical data" view, the left button in the toolbar above the database explorer.
Right-click on the TutorialFem folder > New subject > Subject01
- Keep the default options you set for the protocol.
Right-click on the subject node > Import MRI:
Set the file format: All MRI files (subject space)
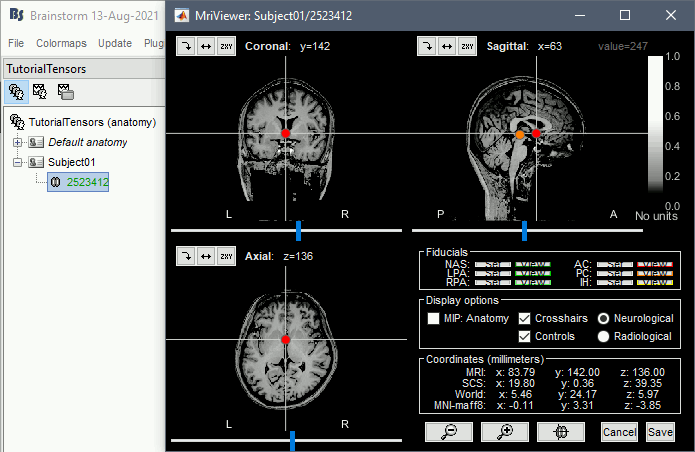
Select the T1 file: BrainSuiteTutorialSVReg/2523412.nii.gz
Click on the link "Click here to compute MNI normalization": option "maff8". This estimates an affine transformation to the MNI space and sets default positions for the anatomical fiducials. The NAS/LPA/RPA fiducials are needed for defining the Brainstorm subject coordinate system, in which the surfaces and FEM meshes are stored.
Diffusion imaging
This computes the This requires BrainSuite to be installed on your computer, with the bdp program available in the system path.
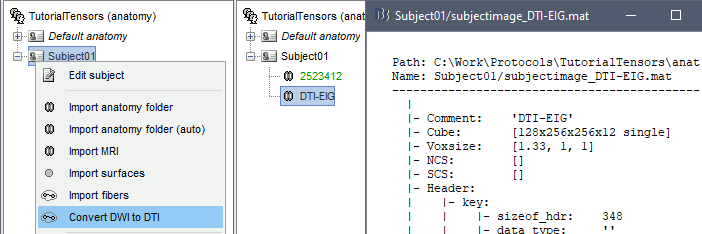
Right-click on Subject01 > Convert DWI to DTI
Select the DWI file: DWI/2523412.dwi.nii.gz
The associated text files *.bvec (orientation of the gradient) and *.bval (value of the gradient) must be in the same folder, with the same file name. Theses files are created from for the DWI acquisition. If you don't have them, ask the person who programmed your DWI sequence and get the files that are specific to your use case.
The process can take up to 30min. At the end, a new file DTI-EIG appears in the database (DTI=diffusion tensors images, EIG=eigenvalue). This file contains 12 volumes, ie. 12 values for each voxel. From 1 to 9: components of the three eigenvectors; from 10 to 12: the values of their norm to the eigenvalue.
FEM mesh
The FEM approach requires a segmentation of the head volume in different tissues, represented as hexahedral or tetrahedral 3D meshes. The methods available within Brainstorm are listed in the tutorial FEM mesh generation.
Here we illustrate only the use of Brain2mesh: this is not the most accurate solution for MRI segmentation but it is probably the fastest solution to obtain a tetrahedral mesh of the head with 5 tissues (gray matter, white matter, CSF, skull, skin). For more accurate results, we recommend using SimNIBS with T1+T2, as illustrated in the tutorial FEM median nerve example.
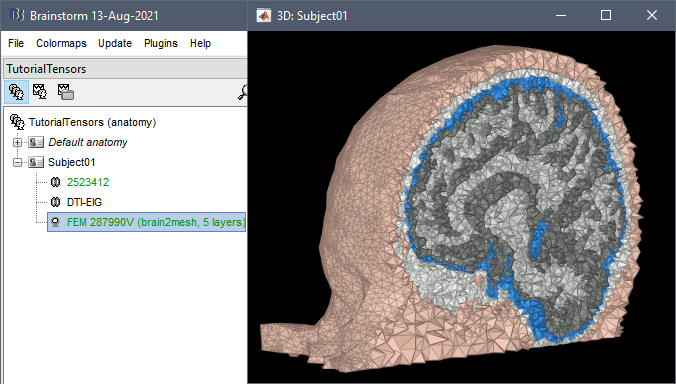
Right-click on the T1 MRI > MRI segmentation > Generate FEM mesh > Brain2mesh.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
After less than 15 minutes, you will obtain a new FEM mesh in the database.
Conductivity tensor generation from DTI
The Effective Medium Approach is applied to convert the diffusion tensors to the conductivity tensors.
www.pnas.org/content/98/20/11697
Computation of FEM mesh tensors
Once the FEM mesh and the DTI tensors are available in the Brainstorm database, the next step for the FEM tensors can be performed by the following:
- Right-click on the FEM mesh - Compute FEM tensors
Brainstorm checks the available tissues in the FEM head model and displays the following panel
This panel lists the tissues available in the FEM head model and assigns a default value of the conductivity for each compartment. Users can change these values to their own if needed.
DTI values can be used to generate conductivity tensors for the white matter (and in some cases for the grey matter). Please, note that the DWI can be used only for the brain tissues and not for the outers compartments (skull and skin)
In this tutorial (and in most cases) we select the white matter. Select the WM anisotropy and keep all the other tissues as isotropic, then these additional options appear asking for the method to use.
The available methods are:
- Effective Medium approach (EMA)
- Effective Medium approach with volume constraints (EMA + VC)
- Simulated or the artificial anisotropy
Only the two first methods require the DTI. More information about these methods can be found on these references [ref1][ref2] and in our main paper [link]
In this tutorial, we use the method "EMA + VC", where the final tensors are constrained to fits the volume of the equivalent isotropic tensor volume.
Visulation of FEM mesh tensors
Once the FEM tensors are successfully computed, they are stored in the FEM head node. By right-clicking on the FEM head, new menu items are added that gives the possibilities to display the FEM tensors either as ellipsoids or as vectors in the direction of the main eigenvector.
The tensors can be displayed either on the FEM mesh or overlaid on the MRI. The following figures show an example of the obtained tensors displayed on the white matter.
On the left, the tensors as a line on the direction of the main eigenvector. On the right, the tensors displayed as ellipsoids. The orientation of the tensor is color-coded as follows: red for right-left, green for anterior-posterior, and blue for superior-inferior.
Note that the quality of the tensors depends on the DWI data and the number of acquisition direction.
Users can also display the tensors on specific tissues, for example on the white matter (left figure) or overlay on the MRI (right figure).
Recommendation
In the case where the user wants to use generate isotropic tensors, then the DTI is not required. For that case, keep all the options to 'isotropic', the recommended display is the 'Ellipsoids', and the final shape will be a sphere (isotropic direction). In the case where more than one FEM head model is in the database, the highlighted one in the green color will be used.
Simulated conductivity tensor
In the case where the DWI is not available, or in the case where the users desire to evaluate the effect of the conductivity change on the head model, the artificial conductivity can be used.
Users can reach this option by following this tutorial and select the third method in this panel. ![]()
Two approaches are integrated within Brainstorm. Either Wang's constraint or the volume's constraint (Wolters). The common feature between these methods is the ratio between the transversal and longitudinal conductivity ratio.
A common example is the skull anisotropy simulation, where the longitudinal conductivity can be higher than the transversal conductivity, the ratio can vary from 2 to 10 [ref]. In this tutorial, we keep all the tissue as isotropic, except the skull, we use a ratio of 0.1 and select the volume constraint. The following figures show the results of this example.
"eigenvalues parallel (longitudinal) and perpendicular (transverse) to the fiber directions" for 1:10 anisotropy (transverse:longitudinal)
Troubleshooting
To be completed soon and linked to BrainSuite website
References
===TODO===
Check the error in the simnibs mesh in X direction and overlay on mri check the error with the brain2mesh Correct the ratio from integer to float check the meaning of transversal/longitidunal in the code add an interactive way yo change the size of the tensor.. important correct the name of the simulated method, correct the EMC and remove the VC and change the coefficcient
