|
Size: 25984
Comment:
|
Size: 30301
Comment: links and small notes about PCA, and scouts through processes
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 5: | Line 5: |
Here, all the steps are done from the scout panel, but scout time series can also be saved with the process "Extract > Scout time series" for further analysis, or computed directly within various processes, e.g. many under Frequency or Connectivity. |
|
| Line 15: | Line 17: |
| We will start by creating regions of interest corresponding to the auditory cortices to illustrate the tools, then define other regions to explore better the dynamics of the brain response. | We will start by creating regions of interest corresponding to the auditory cortices to illustrate the tools, then define other regions to better explore the dynamics of the brain response. |
| Line 18: | Line 20: |
| Almost all the features related to the scouts manipulation are accessible in the ''Scout'' tab in the main Brainstorm window. The scouts are '''automatically saved''' in the surface file from which they are created, and they are loaded and automatically displayed each time the surface is loaded. An '''atlas '''designates in this context a list of scouts. For one cortex surface, we can have as many atlases as needed. An atlas can be an anatomical parcellation (like the ones loaded when using FreeSurfer), a random parcellation generated by Brainstorm, or a user-defined list of ROIs. All the surfaces contain by default an empty atlas called "User scouts", for the user to create new regions of interest. |
Almost all the features related to scout manipulation are accessible in the ''Scout'' tab in the main Brainstorm window. The scouts are '''automatically saved''' in the surface file from which they are created, and they are loaded and automatically displayed each time the surface is loaded. An '''atlas '''designates, in this context, a list of scouts. For one cortex surface, we can have as many atlases as needed. An atlas can be an anatomical parcellation (like the ones loaded when using FreeSurfer), a random parcellation generated by Brainstorm, or a user-defined list of ROIs. All the surfaces contain, by default, an empty atlas called "User scouts", for the user to create new regions of interest. |
| Line 23: | Line 25: |
| * In ''Intra-subject'', double-click on the '''standardized deviant''' average.<<BR>>In Run#01, open the average recordings for the deviant condition, to have a time reference. <<BR>>Go to the first peak in the deviant response, around 90ms.<<BR>>In the Surface tab, increase the amplitude threshold to see something relatively focal. The response is larger in the left hemisphere, so let's start with the left.<<BR>><<BR>> {{attachment:open_files.gif||height="146",width="649"}} | * In ''Intra-subject'', double-click on the '''normalized standard '''average.<<BR>>Open the average recordings for the standard condition, to have a time reference. <<BR>>Go to the first peak in the response, around 90ms.<<BR>>In the Surface tab, increase the amplitude threshold to see something relatively focal. The response is larger in the left hemisphere, so let's start with the left.<<BR>><<BR>> {{attachment:open_files.gif||height="138",width="613"}} |
| Line 42: | Line 44: |
| Grow the scout A1L to '''40 vertices''', not in constrained mode. You can read the number of vertices and the estimated cortical area just below the [<<] and [>>] buttons. | Grow the scout A1L to '''20 vertices''', not in constrained mode. You can read the number of vertices and the estimated cortical area just below the [<<] and [>>] buttons. |
| Line 47: | Line 49: |
| Select the souct in the list, click on the second button in the toolbar '''[Display scouts time series]'''. It displays the signal associated with this region for the entire time window (-100ms to 500ms). <<BR>>We can now observe the early response at 50ms (P50) that was not very obvious before. | Select the scout in the list, click on the second button in the toolbar '''[Display scouts time series]'''. It displays the signal associated with this region for the entire time window (-100ms to 500ms). <<BR>>We can now observe the early response at 50ms (P50) that was not very obvious before. |
| Line 52: | Line 54: |
| In the toolbar on the right side of scouts tab, you can find a list display options. Leave your mouse over it for a few seconds to get a description of each of them. | In the toolbar on the right side of the scouts list, you can find a list of display options. Leave your mouse over each button for a few seconds to get a short description. |
| Line 65: | Line 67: |
| * {{attachment:buttonMri.gif}} Center MRI on scout (opens the MRI viewer and set the position to the current scout) | * {{attachment:buttonMri.gif}} Center MRI on scout (open the MRI viewer and show the position of the scout's seed) |
| Line 68: | Line 70: |
| We have added '''40 vertices''' the scout '''A1L'''. Because we are computed a source model with unconstrained dipoles orientations, we have three sources per vertex. This region A1L corresponds to '''120 signals'''. | We have extended the scout '''A1L''' to '''20 vertices'''. Because this is a source model with constrained dipoles orientations, we have one source only per vertex. The region A1L corresponds to '''20 signals'''. |
| Line 72: | Line 74: |
| Here is a description of the different options. In the case of unconstrained sources (3 signals for each vertex, one for each orientation), the function is applied separately for each orientation and produces 3 time series instead of one. For more details, see the code of bst_scout_value.m. * '''Mean''': Average all the signals. * '''PCA''': Take the first mode of the PCA decomposition of all the signals. * '''Fast PCA''': Same as ''PCA'' function, but computes the first PCA decomposition based only on a subset of the most powerful signals. It gives very similar results, but its computation is much faster for scouts that group a large number of dipoles (>50). * '''Mean(norm)''': Average the absolute values of all the signals: mean(abs(signal)) * '''Max''': For each time point, get the maximum across all the vertices.<<BR>>Signed maximum: m = max(abs(signal)), scout = sign(signal(m)) * m * '''Power''': Average the square of all the signals: mean(signal^2) * '''All''': Do not apply any operation on the scouts signals, returns all the signals. <<BR>><<BR>> {{attachment:scout_all.gif||height="146",width="447"}} |
Here is a description of the different options. In the case of unconstrained sources (3 signals for each vertex, one for each orientation), the function is applied separately for each orientation and produces 3 time series instead of one. For more details, see the code of '''bst_scout_value.m'''. * '''Mean''': Average all the signals. <<BR>><<BR>> {{attachment:scout_mean.gif||width="400"}} * '''PCA''': Take the first mode of the Principal Component Analysis decomposition of all the signals. Here, this is computed on a single file for display, but PCA works best when all epochs and conditions are processed together, through the process tabs. See [[https://neuroimage.usc.edu/brainstorm/Tutorials/PCA|PCA tutorial]]. * '''Fast PCA''': Same as ''PCA'' function, but computes the first PCA decomposition based only on a subset of the strongest signals. It gives very similar results, but its computation is much faster for scouts that group a large number of dipoles (>50). <<BR>><<BR>> {{attachment:scout_pca.gif||width="400"}} * '''Mean(norm)''': Average the absolute values of all the signals: mean(abs(signal)) <<BR>><<BR>> {{attachment:scout_meannorm.gif||width="400"}} * '''Max''': For each time point, get the maximum across all the vertices.<<BR>>Signed maximum: m = max(abs(signal)), scout = sign(signal(m)) * m <<BR>><<BR>> {{attachment:scout_max.gif||width="400"}} * '''Power''': Average the square of all the signals: mean(signal^2) <<BR>><<BR>> {{attachment:scout_power.gif||width="400"}} * '''All''': Do not apply any operation on the scouts signals, returns all the signals. <<BR>><<BR>> {{attachment:scout_all.gif||width="400"}} Note that the function selected here is ignored when running a process with a scout function option, like "Extract > Scout time series" or Connectivity. |
| Line 83: | Line 87: |
| As said in the previous tutorial, the minimum norm current amplitudes can positive or negative depending on the dipole orientation. This means that the values of the scouts, depending on the function that is used, may be positive or negative too. Most of the time we are interested in visualizing the absolute values of the scouts time series, to compare the activation level in different conditions or subjects. For this purpose, at the bottom of the Scout tab, you can choose to display either ''Absolute ''or ''Relative ''values. The effect of this options depends if you are processing sources files with constrained (1 signal per vertex) or unconstrained (3 signals per vertex) dipoles orientations: * '''Constrained''': Apply the scout function to all source signals, then: * '''Absolute''': abs(ScoutFunc(sources)) * '''Relative''': ScoutFunc(sources) * '''Unconstrained''': Apply the scout function to the source signals for each orientation (x,y,z) separately, and then returns either the norm of the 3 orientations, or each orientation separately: * '''Absolute''': Returns __one__ time series: <<BR>>sqrt( ScoutFunc(sources_x).<<HTML(^)>>2 + ScoutFunc(sources_y).<<HTML(^)>>2 + ScoutFunc(sources_z).<<HTML(^)>>2) * '''Relative''': Returns __three__ time series: <<BR>>ScoutFunc(sources_x), ScoutFunc(sources_y), ScoutFunc(sources_z) '''Display only''': Note that it is a display option, it is not saved in the scouts themselves. It is used only when displaying the scouts time series with the [Display scouts time series] button of the Scout tab. In all other cases, like the extraction of the scouts values from a script, this relative/absolute option is ignored. == Relative Z-score values == Change the selection to '''Relative '''and display again the activation of scout A1L for the deviant condition. It shows only one signal because the three orientations were already grouped in the one value during the '''Z-score normalization'''. The sign of the Z-score is important to look at, because it indicates if the values are higher or lower than the baseline. With this representation, we can confirm that we only observe higher activity values than during the baseline. . {{attachment:scout_relative.gif||height="155",width="504"}} Select again the '''Absolute''' display again, and keep it for the rest of the tutorial. |
As said in the previous tutorial, the minimum norm current amplitudes can be positive or negative depending on the dipole orientation. This means that the values of the scouts, depending on the function that is used, may be positive or negative too. Most of the time we are interested in visualizing the absolute values of the scouts time series, to compare the activation level in different conditions or subjects. But it is sometimes easier to understand the temporal dynamic of a ROI with the relative values. At the bottom of the Scout tab, you can choose to display either ''Absolute ''or ''Relative ''values. <<BR>>The effect of this options changes whether you are processing source files with constrained (1 signal per vertex) or unconstrained (3 signals per vertex) dipoles orientations: '''Constrained''': Apply the scout function to all source signals, then: * '''Absolute''': abs(ScoutFunc(sources))<<BR>><<BR>> {{attachment:scout_absolute.gif||height="144",width="465"}} * '''Relative''': ScoutFunc(sources)<<BR>><<BR>> {{attachment:scout_relative.gif||height="144",width="466"}} '''Unconstrained''': Apply the scout function to the source signals for each orientation (Sx,Sy,Sz) separately, and then returns either the norm of the 3 orientations, or each orientation separately (except PCA, see below): * '''Absolute''': Returns __one__ time series: <<BR>>sqrt( ScoutFunc(Sx)^2^ + ScoutFunc(Sy)^2^ + ScoutFunc(Sz)^2^) <<BR>><<BR>> {{attachment:scout_unconstr_abs.gif||height="147",width="466"}} * '''Relative''': Returns __three__ time series: <<BR>>A1L1=ScoutFunc(Sx), A1L2=ScoutFunc(Sy), A1L3=ScoutFunc(Sz) <<BR>><<BR>> {{attachment:scout_unconstr_rel.gif||height="148",width="466"}} * Since PCA can also be used to combine the 3 orientations, it is applied here across all sources and orientations at once, producing a single time series, like in the constrained case above. '''Display only''': Note that this relative/absolute selection is a display option, it is not saved in the scouts themselves. It is used only when displaying the scouts time series with the [Display scouts time series] button of the Scout tab. In all other cases, such as the extraction of the scouts values from a script, this relative/absolute option is ignored. In those cases, there may also be an option to [[https://neuroimage.usc.edu/brainstorm/Tutorials/PCA#Unconstrained_source_flattening_with_PCA|f]][[https://neuroimage.usc.edu/brainstorm/Tutorials/PCA#Unconstrained_source_flattening_with_PCA|latten unconstrained source orientations]] first, resulting in a single time series per scout instead of 3. |
| Line 107: | Line 108: |
| * Select the scout '''A1L '''and click on ['''Display soutces time series'''].<<BR>><<BR>> {{attachment:scout_nooverlay.gif||height="126",width="451"}} * In the Record tab, you can use the button '''[=]''' to configure the amplitude of multiple axes. <<BR>>Select it to force multiple graphs to use the same scale, unselect it to scale each graph separately. <<BR>> You can find the same option in the figure popup menu: '''Figure > Uniform figure scale'''. <<BR>><<BR>> {{attachment:scout_equalamp.gif||height="126",width="450"}} * At the bottom of the Scout tab, select the option "'''Overlay: Conditions'''", then click [Display] again.<<BR>><<BR>> {{attachment:scout_overlay_cond.gif||height="143",width="607"}} * The Z-score value for the standard condition is a lot higher than the deviant condition because of the number of trials we used for computing the two averages. The SNR is higher with more trials, the baseline has less variance and the Z-score can be higher. |
* Select the scout '''A1L '''and click on ['''Display scouts time series''']. <<BR>>This computes the scouts time series for all the files that are currently open.<<BR>><<BR>> {{attachment:scout_nooverlay2.gif||height="122",width="436"}} * In the Record tab, you can use the button '''[=]''' to configure the amplitude of multiple axes. <<BR>>Select it to force multiple graphs to use the same scale, unselect it to scale each graph separately. <<BR>> You can find the same option in the figure popup menu: '''Figure > Uniform figure scale'''. <<BR>><<BR>> {{attachment:scout_nooverlay.gif||height="121",width="435"}} * At the bottom of the Scout tab, select the option "'''Overlay: Files'''", then click [Display] again.<<BR>><<BR>> {{attachment:scout_overlay_cond.gif||height="157",width="573"}} * The '''Z-score value''' for the '''standard condition is a lot higher''' than the deviant condition because of the number of trials we used for computing the two averages (5x more trials for the standard). The SNR is higher with more trials, the baseline has less variance so the Z-score is higher. * To overlay the results of two conditions averaged with very different numbers of trials, it makes more sense to display the scout time series for non-normalized maps (averaged current density).<<BR>><<BR>> {{attachment:scout_overlay_cond2.gif||height="159",width="575"}} |
| Line 115: | Line 118: |
| * In the Surface tab, smooth the cortical surface at '''60%'''. | * In the Surface tab, smooth the cortical surface at '''70%'''. |
| Line 117: | Line 120: |
| * Grow all the regions to the same size: '''40 vertices'''. * In the screen captures below, the scout times series have been computed using the '''non-normalized''' files, in order to plot the standard and deviant averages in the same graph. * '''A1L''': Left primary auditory cortex (Heschl gyrus) - Already marked. |
* Grow all the regions to the same size: '''20 vertices'''. * '''A1L''': Left primary auditory cortex (Heschl gyrus) - __Already marked__. |
| Line 121: | Line 123: |
| * '''Deviant '''condition, t='''90ms''', amplitude threshold='''60%''' <<BR>><<BR>> {{attachment:scout_a1l.gif}} | * '''Standard '''condition, t='''90ms''', amplitude threshold='''50%''' <<BR>><<BR>> {{attachment:scout_a1l_constr.gif||height="139",width="559"}} |
| Line 123: | Line 125: |
| * The response in this region is a lot less obvious than A1L. These binaural auditory stimulations should be generating similar responses in both left and right auditory cortices at early latencies. Possible explanations for this observation: | * The response in this region is not as obvious than A1L. These binaural auditory stimulations should be generating similar responses in both left and right auditory cortices at early latencies. Possible explanations for this observation: |
| Line 127: | Line 129: |
| * The orientation of the source makes it difficult for the MEG sensors to capture. * '''Deviant '''condition, t='''90ms''', amplitude threshold='''40% '''<<BR>><<BR>> {{attachment:scout_a1r.gif}} |
* The orientation of the source makes it more difficult for the MEG sensors to capture. * '''Deviant '''condition, t='''90ms''', amplitude threshold='''50% '''<<BR>><<BR>> {{attachment:scout_a1r_constr.gif||height="140",width="560"}} |
| Line 132: | Line 134: |
| * '''Deviant '''condition, t='''130ms''', amplitude threshold='''30% '''<<BR>><<BR>> {{attachment:scout_igfl.gif}} | * '''Deviant '''condition, t='''130ms''', amplitude threshold='''30% '''<<BR>><<BR>> {{attachment:scout_ifgl_constr.gif||height="140",width="560"}} |
| Line 137: | Line 139: |
| * At 240ms, the peak in the standard condition is probably a motor preparation. At 350ms, the motor task begins, the subject moves the right hand (recorded reaction times 500ms +/- 108ms). | * At 230ms, the peak in the deviant condition is probably a motor preparation. At 350ms, the motor task begins, the subject moves the right hand (recorded reaction times 500ms +/- 108ms). |
| Line 139: | Line 141: |
| * '''Standard '''condition, t='''170ms''', amplitude threshold='''60%''' <<BR>><<BR>> {{attachment:scout_m1l.gif}} * '''SMAL''': Left supplementary motor area * Probably involved in the preparation and control of the motor response as well. * '''Deviant '''condition, t='''440ms''', amplitude threshold='''40%'''''' '''<<BR>><<BR>> {{attachment:scout_smal.gif}} * '''PPCR''': Right posterior parietal cortex * Known to play a role as a relay in the auditory processing. * '''Standard '''condition, t='''250ms''', amplitude threshold='''25%'''''' '''<<BR>><<BR>> {{attachment:scout_ppcr.gif}} |
* '''Deviant '''condition, t='''440ms''', amplitude threshold='''25%''' <<BR>><<BR>> {{attachment:scout_m1l_constr.gif||height="140",width="560"}} |
| Line 150: | Line 146: |
| We are going to create another scout for the other condition: the stimulation of the '''left thumb'''. 1. Close all figures (toolbar button). 1. Display the times series for ''Subject01 / '''Left '''/ ERF'' (double-click). 1. Display the associated sources (Ctrl+S). 1. Go to the main response peak (46 ms). 1. Adjust the threshold value to get only a very focal point (about 60%). 1. Create a new scout at this point. Name it ''RightSS'', for Right SomatoSensory cortex. 1. Grow it to the same number of vertices as the ''LeftSS ''scout (about 15 vertices). 1. Select both scouts (Ctrl+click) and click on ''Display scouts time series'' button (second button in toolbar).<<BR>><<BR>> {{attachment:scouts2.gif}} <<BR>><<BR>> {{attachment:scoutsTS_LR.gif}} 1. You can display the values of the two scouts on the same graph. Check the ''Overlay > Scouts'' checkbox in the Scout tab, and hit the ''Display scouts time series'' button again.<<BR>><<BR>> {{attachment:scoutsTS_LR_overlay.gif}} 1. You can observe something that we expected: the activity in the '''right somatosensory cortex''' is much higher than in the left somatosensory cortex for a stimulation of the '''left thumb'''. == Display scouts from the database explorer == You have to display the sources on the cortex to create the scouts. But once they are created, you can directly display the scouts time series from the tree. It means that you can quickly compare the values for a scout between many different conditions without having to open them all. 1. Close all the figures (button in toolbar) 1. Select the options "values: absolute" and "overlay: scouts" 1. Right-click on '''Right '''/ ERF / MN > Cortical activations > Scouts time series.<<BR>>By default it shows all the scouts that are available in the current atlas.<<BR>><<BR>> {{attachment:treeRight.gif}} <<BR>> {{attachment:treeRight_TS.gif}} 1. Note that if a surface is loaded, and at least one scout selected in the ''Scout'' tab, this popup menu would display only the selected scouts. 1. You can also display the scout values for many source files at the same time. Now keep the previous figure opened, so you have access to the scouts list in the ''Scout'' tab. Select only the ''LeftSS'' scout. In the database explorer, select at the same time the source files from condition Left and Right > right-click > Cortical activations > Scouts time series.<<BR>><<BR>> {{attachment:treeLR.gif}} <<BR>> {{attachment:treeLR_TS.gif}} 1. You can superimpose the two traces from the two experimental conditions by selecting the ''Overlay > Conditions'' checkbox in ''Scout ''panel. Select again the two files > Right click > Scouts time series. We see now the activation of the LeftSS scout for the stimulation of both thumbs.<<BR>><<BR>> {{attachment:panelOverlayCond.gif}} 1. This menu "Scouts time series" is present at all the levels in the tree. Try right-clicking on Subject01 > Scouts time series. 1. If you select a node in the database tree and click on the ''Display scouts time series'' button in the ''Scout ''tab, it would have exactly the same effect. <<BR>>Try selecting '''both scouts''' in the list, select '''Subject01 '''in the tree, overlay / conditions, and then click on this button. You will see the response for each scout in both conditions.<<BR>><<BR>> {{attachment:scoutsOverlayBoth.gif}} |
We can display the activity of multiple regions simultaneously. * Close everything (toolbar button [X]). * Open the two '''normalized average '''source files (standard and deviant). * In the Scout tab, select the '''A1L '''and '''IFGL '''scouts simultaneously. * Select the option "'''Overlay:Scouts'''", do not select "Overlay:Files", Absolute values. * Click on ['''Display scouts time series'''].<<BR>><<BR>> {{attachment:overlay_scouts.gif||height="142",width="490"}} * Now select the option "'''Overlay:Files'''", do not select "Overlay:Scouts", click on ['''Display''']. <<BR>><<BR>> {{attachment:overlay_conditions.gif||height="143",width="491"}} * In both conditions, we observe similar early responses (P50 and N100), then it diverges.<<BR>>In the deviant condition, we observe a pattern A1-IFG-A1 that is absent in the standard condition. <<TAG(Advanced)>> == From the database explorer == You have to display the sources on the cortex to create the scouts. But once they are created, you can directly display the scouts time series from the database explorer. It means that you can quickly compare the values for a scout between many different conditions without having to open them all. * Close everything (toolbar button [X]). * Select the option "'''Overlay:Files'''", do not select "Overlay:Scouts". * Select the two average recordings in Run#01 folder > right-click > '''Scouts time series'''. <<BR>>Note that this menu is present at all the levels in the database explorer.<<BR>><<BR>> {{attachment:scout_tree_popup.gif||height="223",width="449"}} * If no scout is currently loaded in the Scout tab, it shows all the scouts available in the surface. <<BR>>If a surface is loaded, and at least one scout selected in the Scout tab, this popup menu would display only the selected scouts. <<BR>><<BR>> {{attachment:scout_tree.gif||height="202",width="457"}} * Once the list of scouts is loaded in the Scout tab, you can select one of them, and then display its values for all the trials of a single condition (overlay:files). <<BR>><<BR>> {{attachment:scout_tree_trials.gif||height="218",width="640"}} <<TAG(Advanced)>> == Sign flip == In the case of source models with '''constrained orientations''' (normal to the cortex), the sign of the current can be an issue. If the region is large enough to include vertices with normals in opposite directions, averaging the source values may cancel out the activity. Let's use the example from the previous tutorial and consider that one scout contains all the dipoles corresponding to both the red arrows (positive values) and the blue arrows (negative values). If we average all the values from this scout, we get a value close to zero. . {{attachment:minnorm_sketch.gif||height="146",width="439"}} To avoid this, a mechanism was added in the scout calculation, to flip the sign of sources with opposite directions before the averaging. We start by finding the dominant orientation of the scout, then flip the sign of the values that are not in the same direction (scalar product of the orientations < 0). If the sign of some sources is flipped, you get a message in the Matlab command window, for example:<<BR>>''BST> Flipped the sign of 7 sources.'' |
| Line 179: | Line 184: |
| Let's spend a few minutes to explore all the menus in the ''Scout ''tab. Don't hesitate to click on all of them, it's the best way to remember what they do. |
|
| Line 182: | Line 185: |
| . {{attachment:menuNew.gif}} * '''New atlas''': Create a new empty atlas. |
. {{attachment:menu_atlas.gif}} * '''New atlas''': * '''Empty atlas''': Create a new empty atlas. * '''Copy atlas''': Duplicate an atlas and all the scouts it contains. * '''Copy selected scouts''': Create a new atlas and copy only the selected scouts to it. * '''Source model options''': Create a special atlas "Source model", with which you can attribute different modeling constraints to each region, after merging the cortex with some subcortical regions. This is required to use the option "'''Custom head model'''" (mixed models). * '''Volume scouts''': Create a volume atlas in order to define 3D scouts ([[https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource|volume head models]]). * '''From subject anatomy''': Import as surface or [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource|volume]] scouts the ROIs defined in one of the volume parcellation available in the subject's anatomy ([[https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#MNI_parcellations|MNI-based atlases]] or subject parcellations from [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12#Volume_parcellations|CAT12]] or [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Volume_parcellations|FreeSurfer]]). |
| Line 188: | Line 202: |
| * '''Rename atlas''': Change the name that appears in the atlas list to refer to the selected atlas * '''Delete atlas''': Delete the atlas and all the scouts in contains. The "User scouts" entry cannot be deleted. |
* '''Rename atlas''': Change the name that appears in the atlas list to refer to the selected atlas. * '''Delete atlas''': Delete the current atlas and all the scouts in contains. |
| Line 194: | Line 208: |
| * '''Copy atlas''': Duplicates an atlas and all the scouts * '''Copy selected scouts''': Copy the selected scouts, and paste them in a new empty atlas. * '''Subdivide atlas''': Splits all the scouts of the current atlas in smaller ROIs. Available options:<<BR>><<BR>> {{attachment:scoutSubdivide.gif}} |
* '''Subdivide atlas''': Splits all the scouts of the current atlas in smaller ROIs. Available options:<<BR>><<BR>> {{attachment:scoutSubdivide.gif||height="146",width="298"}} |
| Line 202: | Line 212: |
| * '''Surface clustering''': Create a parcellation of the cortex surface and saves it as a new atlas. Can be useful for studying the connectivity between different regions of the brain. Only the "Random" clustering is currently available. Experimental features. | * '''Surface clustering''': Create a parcellation of the cortex surface and saves it as a new atlas. <<BR>>Only the "Homogeneous parcellation" clustering is currently available. * '''Save modifications''': Force the current modifications to be saved to the cortex surface. * '''Undo all modifications''': Restore the atlas the way it was the last time the surface was loaded. |
| Line 205: | Line 219: |
| . {{attachment:menuEdit.gif}} * '''Add vertices''': The user can select some points on the cortex surface and add them to the scout.<<BR>>Equivalent: Click on ''Select point'' (first toolbar button), then select the scout in list, and select 3d points. * '''Edit in MRI''': Open an interface to edit the select scout as a 3D ROI, slice by slice. * Define the 3D mask using the drawing tools (read the tooltips of the buttons for help). * Adjust the MRI contrast with right-click+move on the MRI image. * Move through the slices with the mouse wheel, the small image on the left, or the slider. * __Warning__: Only the vertices contained in the 3D mask are kept, all the volume information is lost. It means that the mask you intendend to draw might be very different from what you get as a scout at the end...<<BR>><<BR>> {{attachment:menuEditMriMask.gif}} |
. {{attachment:menu_scout.gif}} * '''New: coordinates''': Create a new scout starting from the vertex that is the closest to the specified coordinates. You can enter coordinates in MRI, SCS or MNI coordinates. * '''Add vertices''': The user can select some points on the cortex surface and add them to the scout.<<BR>>Equivalent: Click on ''Select point'' (first toolbar button) then select the scout in list. * '''Edit in MRI''': Open an interface to edit the selected scout as a 3D ROI, slice by slice. Only the vertices contained in the 3D mask are kept, all the volume information is lost. It means that the mask you intended to draw might be very different from what you get as a scout at the end. This is not a very reliable tool, as there is no direct correspondence between the volume and the surface. |
| Line 217: | Line 229: |
| * '''Set region''': Set the cortical region for the selected scouts. This information is useful when representing functional connectivity graphs. * '''Rename''': Rename the selected scout. Shortcut: ''double-click''. * '''Set color''': Change the display color of scout patch in 3D figures. * '''Delete''': Delete selected scouts. Shortcut: ''Delete'' key |
* '''Set region''': Set the cortical region for the selected scouts. * '''Rename''': Rename the selected scout. Shortcut: '''double-click'''. * '''Set color''': Change the display color of the selected scout. * '''Delete''': Delete selected scouts. Shortcut: '''Delete '''key |
| Line 223: | Line 235: |
| * '''Export to Matlab''': Export the structures of the selected scouts to the Matlab environment, and makes them accessible from the command window. This menu can be useful to get quickly the list of vertices in the scout. | * '''Difference''': With two scouts A and B selected, this menu removes the vertices included scouts A from scout B, or vice versa. The direction of the difference is asked in a dialog window. * '''Export to Matlab''': Export the structures of the selected scouts to the Matlab environment and makes them accessible from the command window. This menu can be useful to get quickly the list of vertex indices or modify a scout manually. |
| Line 225: | Line 238: |
| * '''Project to''': Project the selected scout on another surface available in the database. * '''Project to: Contralateral hemisphere''': Transfer scouts defined on one hemisphere to the other hemisphere. This feature is only available with FreeSurfer, and the recon-all segmentation pipeline must be executed with [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Run_FreeSurfer_manually|additional options]]. In the tutorial dataset and in the ICBM152 template anatomy, this option was not used, and therefore this menu generates an error. To get this contralateral projection to work, FreeSurfer recon-all must be executed again with the extra option, and the anatomy of the subject must be imported again. [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Contralateral_registration|More information]]. |
|
| Line 228: | Line 243: |
| . {{attachment:menuView.gif}} * '''Simulate recordings''': Multiply the selected scouts with the forward model. Simulate the scalp data that would be recorded if only the selected cortex region was activated; all the other sources are set to zero. Create a new data file in the database.<<BR>>If no scout selected: simulate recordings produced by the activity of the entire cortex. |
. {{attachment:menu_sources.gif}} |
| Line 231: | Line 245: |
| * '''Expand with correlation''': Computes the correlation between the values for the scout's seed (first point) and all the other sources. The sources that have correlation coefficient superior to a given threshold are added to the scout. Correlation is computed with the Matlab ''corrcoef ''function. | * '''Expand with correlation''': Computes the correlation between the values for the scout's seed (first point) and all the other sources. The sources that have correlation coefficient superior to a given threshold are added to the scout. |
| Line 234: | Line 248: |
| * '''Simulate recordings''': Multiply the selected scouts with the forward model. Simulate the scalp data that would be recorded if only the selected cortex region was activated; all the other sources are set to zero. Create a new data file in the database.<<BR>>If no scout selected: simulate recordings produced by the activity of the entire cortex. | |
| Line 242: | Line 257: |
| The region for a scout can be set with the ''Scout > Set region'' menus, and is encoded in a string that contains at least 2 characters: "'''HLxxx'''". H represents the hemisphere (L,R,U), L stands for the lobe (F,PF,C,P,T,O,L,U), and xxx for the sub-region name (optional). For both the hemisphere and the lobe, the value "U" stands for "Undefined", meaning that the classification is simply not set. The menu ''Set region>Custom region...'' lets you edit this string directly. The menu ''Set region>Undefined ''sets the region string to "UU" (undefined hemisphere, undefined lobe). When set, the region string is shown before the scout name in the list, representing only the defined levels. It doesn't show the letters U for "undefined", therefore the scouts you create manually, that are by default set to "UU", do not show any extra string. . {{attachment:menuRegions.gif}} <<TAG(Advanced)>> == Unconstrained sources == |
The region for a scout can be set with the ''Scout > Set region'' menus, and is encoded in a string that contains at least 2 characters: "'''HLxxx'''". H represents the hemisphere (L,R,U), L stands for the lobe (F,PF,C,P,T,O,L,U), and xxx for the sub-region name (optional). For both the hemisphere and the lobe, the value "U" stands for "Undefined", meaning that the classification is simply not set. The menu ''Set region>Custom region...'' lets you directly edit this string. When set, the region string is shown before the scout name in the list, representing only the defined levels. It doesn't show the letters U for "undefined". . {{attachment:scout_region.gif}} |
| Line 254: | Line 266: |
| The scouts are saved in the surface file on which they have been defined.<<BR>> In the anatomy view, right-click on the selected cortex surface (cortex_15002V) > View file contents. {{attachment:scout_harddrive.gif||height="431",width="569"}} '''iAtlas''': Index of the atlas that is currently selected for this surface. '''Atlas''': Array of structures, each entry is one menu in the drop-down list in the Scout tab. * '''Name''': Label of the atlas (reserved names: "User scouts", "Structures", "Source model"). * '''Scouts''': Array of structures, one per scout in this atlas. * '''Vertices''': Array of indices of the vertices that are part of the scout. * '''Seed''': Index of the central point of the scout (or the most relevant one). * '''Color''': [r,b,g] color array, with values between 0 and 1. * '''Label''': Display name of the scout (must be unique in this atlas). * '''Function''': Scout function {'Mean', 'PCA', 'FastPCA', 'Mean_norm', 'Max', 'Power', 'All'} * '''Region''': Code name for indicating the anatomical region in which the scout is located. * '''Handles''': Graphic handles if the scout is currently displayed (always empty in a file). ==== Useful functions ==== * '''bst_scout_value''': Combine multiple signals into one. * '''process_extract_scout''': Process "Extract > Scouts time series" * '''view_scouts''': Compute scouts time series and displays them in a new figure. == Additional documentation == * Forum: Scout coordinates: http://neuroimage.usc.edu/forums/showthread.php?2375 * Forum: Coordinates of the max value from a scout: https://neuroimage.usc.edu/forums/t/get-the-coordinates-of-a-max-value-from-scouts/4959/2 * Forum: Importing other FreeSurfer atlases: https://neuroimage.usc.edu/forums/t/930 |
Tutorial 23: Scouts
Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Richard Leahy, Sylvain Baillet
In Brainstorm jargon, a scout represents a region of interest (ROI) in the available source space. It is a subset of dipoles defined on the cortex surface or the head volume. This tutorial explains how to create one or several scouts, use them to represent the activity in specific brain regions and compare the responses between different experimental conditions.
Here, all the steps are done from the scout panel, but scout time series can also be saved with the process "Extract > Scout time series" for further analysis, or computed directly within various processes, e.g. many under Frequency or Connectivity.
Contents
Hypothesis
For all the brain imaging experiments, it is highly recommended to have a clear hypothesis to test before starting the analysis of the recordings. With this auditory oddball experiment, we would like to explore the temporal dynamics of the auditory network, the deviant detection and the motor response. According to the literature, we expect to observe at least the following effects:
Bilateral response in the primary auditory cortex (P50, N100), in both experimental conditions (standard and deviant beeps).
Bilateral activity in the inferior frontal gyrus and the auditory cortex corresponding to the detection of an abnormality (latency: 150-250ms) for the deviant beeps only.
Decision making and motor preparation, for the deviant beeps only (after 300ms).
We will start by creating regions of interest corresponding to the auditory cortices to illustrate the tools, then define other regions to better explore the dynamics of the brain response.
Creating a scout
Almost all the features related to scout manipulation are accessible in the Scout tab in the main Brainstorm window. The scouts are automatically saved in the surface file from which they are created, and they are loaded and automatically displayed each time the surface is loaded.
An atlas designates, in this context, a list of scouts. For one cortex surface, we can have as many atlases as needed. An atlas can be an anatomical parcellation (like the ones loaded when using FreeSurfer), a random parcellation generated by Brainstorm, or a user-defined list of ROIs. All the surfaces contain, by default, an empty atlas called "User scouts", for the user to create new regions of interest.
First vertex (seed)
In Intra-subject, double-click on the normalized standard average.
Open the average recordings for the standard condition, to have a time reference.
Go to the first peak in the response, around 90ms.
In the Surface tab, increase the amplitude threshold to see something relatively focal. The response is larger in the left hemisphere, so let's start with the left.
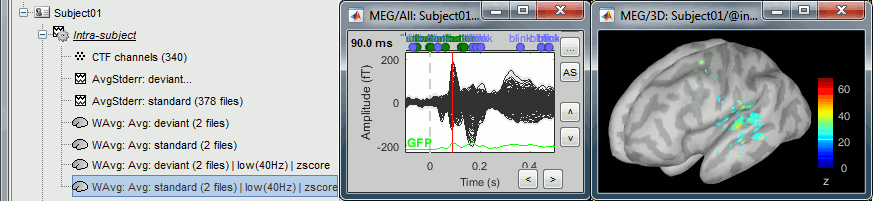
Switch to the Scout tab, click on the first button in the toolbar (the big cross).
In the 3D figure, click where we expect to find the primary auditory cortex (rotate and zoom before if necessary). A small green dot with the label "1" appears where you clicked. Your first scout is created in the User scouts atlas, and contains only one vertex for the moment.
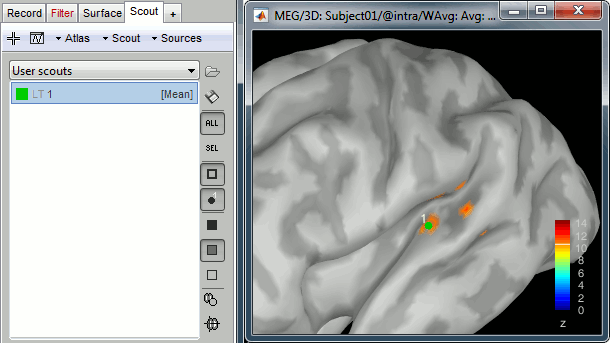
If you are not satisfied with the position of the vertex you selected, delete the new scout (select it in the list and press the Delete key, or menu Scout > Delete) and try again.
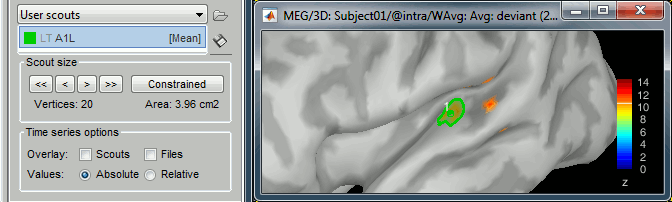
Rename it "A1L", for primary auditory left: double-click on it in the list, or menu Scout > Rename.
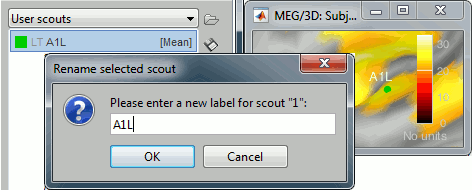
In light grey, you can see in the list the letters "LT", this means that based on the anatomical atlases imported from FreeSurfer, the point you clicked is in the left temporal lobe.
Growing a scout
For now, our scout contains only one vertex of the cortex surface. Most of the time, the aim of a scout is to extract the average activity over a larger region. The buttons in the Scout size section offer basic operations to define the scout extension.
[>]: Add the closest vertex (with respect to the seed) to the current scout.
[<]: Remove the furthest vertex (with respect to the seed) from the current scout.
[>>]: Grow scout in all directions.
[<<]: Shrink scout in all directions.
'Constrained': If this button is pressed, only the vertices that have a source value above the threshold will be added to the scout (its growth will be limited to the colored patch on the surface).
Add vertex manually: Select the Select point button, then select again the "A1L" scout. When you click on a vertex on the surface, it is added to the selected scout.
Remove vertex manually: Same as previously, but holding the SHIFT key at the same time.
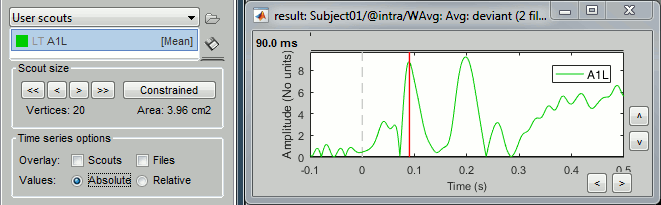
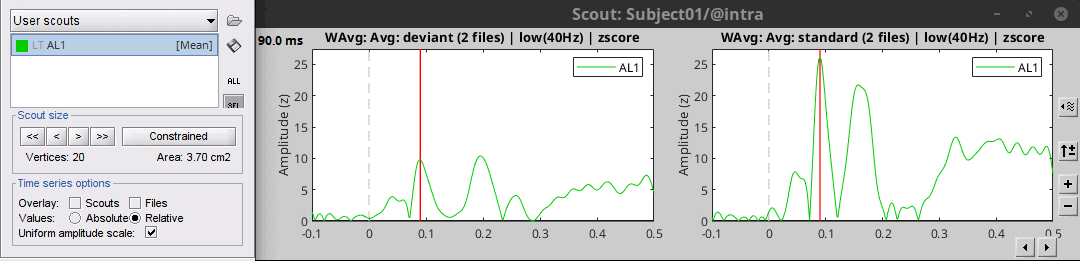
Grow the scout A1L to 20 vertices, not in constrained mode. You can read the number of vertices and the estimated cortical area just below the [<<] and [>>] buttons.
Display time series
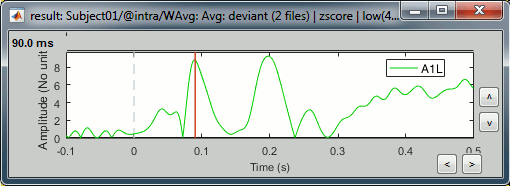
Select the scout in the list, click on the second button in the toolbar [Display scouts time series]. It displays the signal associated with this region for the entire time window (-100ms to 500ms).
We can now observe the early response at 50ms (P50) that was not very obvious before.
3D display options
In the toolbar on the right side of the scouts list, you can find a list of display options. Leave your mouse over each button for a few seconds to get a short description.
 Load Atlas
Load Atlas  Save selected scouts
Save selected scouts  Show all the scouts
Show all the scouts  Show only the selected scouts
Show only the selected scouts Hide all: Uncheck both buttons above to hide the scouts in the 3D figure.
 Show / hide the contour line
Show / hide the contour line  Show / hide the scouts labels
Show / hide the scouts labels  Scout patch: Opaque
Scout patch: Opaque  Scout patch: Transparent
Scout patch: Transparent  Scout patch: None
Scout patch: None  Display the color of the region instead of the scout color (only for anatomical atlases)
Display the color of the region instead of the scout color (only for anatomical atlases)  Center MRI on scout (open the MRI viewer and show the position of the scout's seed)
Center MRI on scout (open the MRI viewer and show the position of the scout's seed)
Scout function
We have extended the scout A1L to 20 vertices. Because this is a source model with constrained dipoles orientations, we have one source only per vertex. The region A1L corresponds to 20 signals.
The signals are grouped together into one unique signal that is then used to represent the activity of the region of interest. In the list of available scouts, you can see the indication [Mean] next to the name of the scout. It represents the name of the function that is used for combining all the source signals into one. This function can be changed individually for each scout, with the menu Scout > Set function.
Here is a description of the different options. In the case of unconstrained sources (3 signals for each vertex, one for each orientation), the function is applied separately for each orientation and produces 3 time series instead of one. For more details, see the code of bst_scout_value.m.
Mean: Average all the signals.
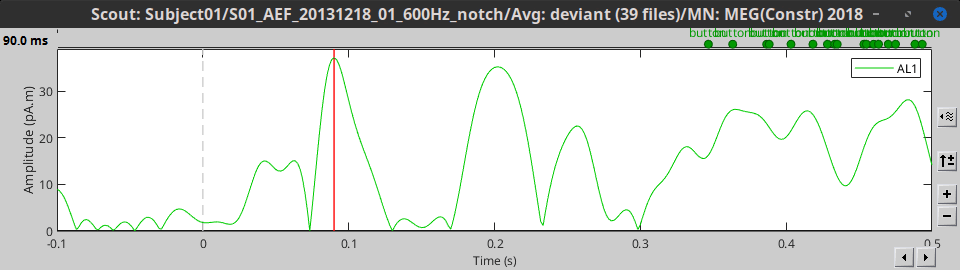
PCA: Take the first mode of the Principal Component Analysis decomposition of all the signals. Here, this is computed on a single file for display, but PCA works best when all epochs and conditions are processed together, through the process tabs. See PCA tutorial.
Fast PCA: Same as PCA function, but computes the first PCA decomposition based only on a subset of the strongest signals. It gives very similar results, but its computation is much faster for scouts that group a large number of dipoles (>50).
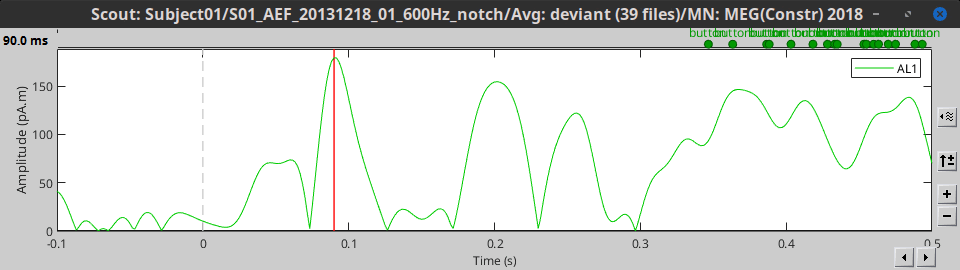
Mean(norm): Average the absolute values of all the signals: mean(abs(signal))
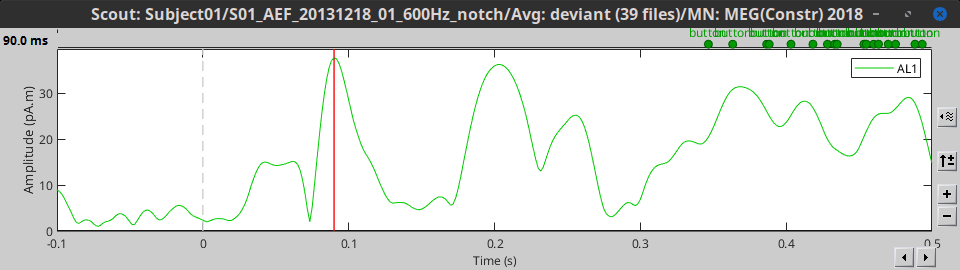
Max: For each time point, get the maximum across all the vertices.
Signed maximum: m = max(abs(signal)), scout = sign(signal(m)) * m
Power: Average the square of all the signals: mean(signal^2)
All: Do not apply any operation on the scouts signals, returns all the signals.
Note that the function selected here is ignored when running a process with a scout function option, like "Extract > Scout time series" or Connectivity.
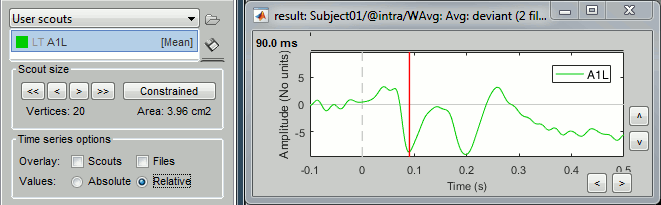
Option: Absolute / relative
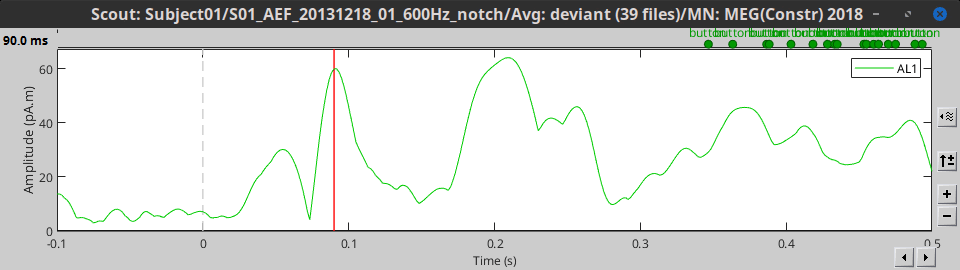
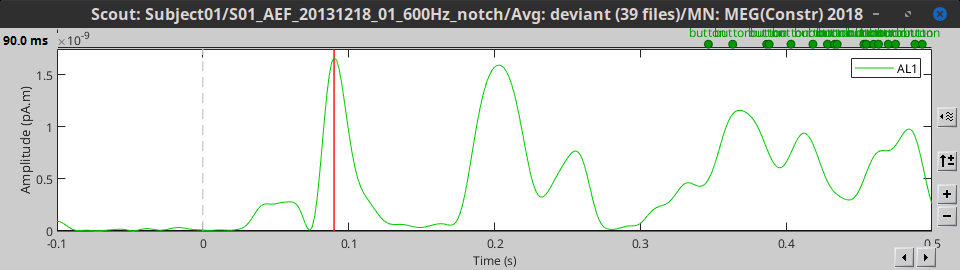
As said in the previous tutorial, the minimum norm current amplitudes can be positive or negative depending on the dipole orientation. This means that the values of the scouts, depending on the function that is used, may be positive or negative too. Most of the time we are interested in visualizing the absolute values of the scouts time series, to compare the activation level in different conditions or subjects. But it is sometimes easier to understand the temporal dynamic of a ROI with the relative values.
At the bottom of the Scout tab, you can choose to display either Absolute or Relative values.
The effect of this options changes whether you are processing source files with constrained (1 signal per vertex) or unconstrained (3 signals per vertex) dipoles orientations:
Constrained: Apply the scout function to all source signals, then:
Absolute: abs(ScoutFunc(sources))
Relative: ScoutFunc(sources)
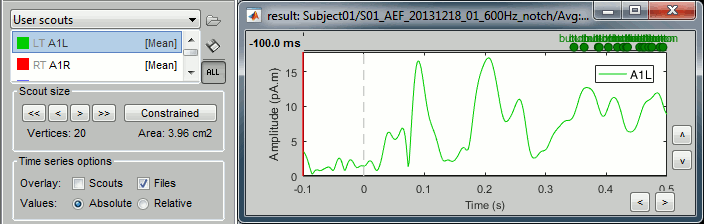
Unconstrained: Apply the scout function to the source signals for each orientation (Sx,Sy,Sz) separately, and then returns either the norm of the 3 orientations, or each orientation separately (except PCA, see below):
Absolute: Returns one time series:
sqrt( ScoutFunc(Sx)2 + ScoutFunc(Sy)2 + ScoutFunc(Sz)2)
Relative: Returns three time series:
A1L1=ScoutFunc(Sx), A1L2=ScoutFunc(Sy), A1L3=ScoutFunc(Sz)
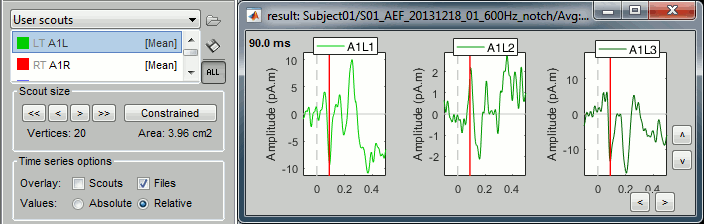
- Since PCA can also be used to combine the 3 orientations, it is applied here across all sources and orientations at once, producing a single time series, like in the constrained case above.
Display only: Note that this relative/absolute selection is a display option, it is not saved in the scouts themselves. It is used only when displaying the scouts time series with the [Display scouts time series] button of the Scout tab. In all other cases, such as the extraction of the scouts values from a script, this relative/absolute option is ignored. In those cases, there may also be an option to flatten unconstrained source orientations first, resulting in a single time series per scout instead of 3.
Multiple conditions
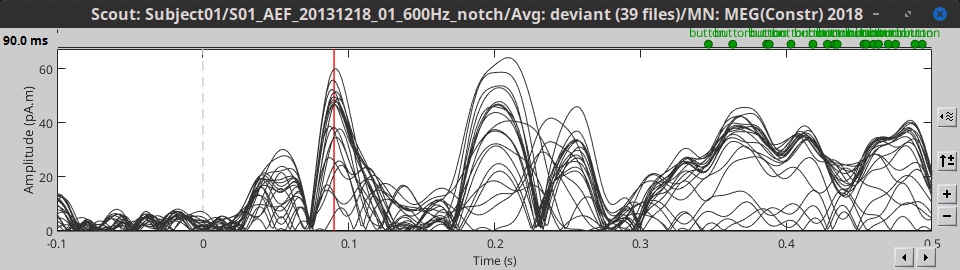
We can easily compare the activity between multiple conditions and multiple regions of interest.
In Intra-subject, open at the same time the normalized sources for deviant and standard condition.
Select the scout A1L and click on [Display scouts time series].
This computes the scouts time series for all the files that are currently open.
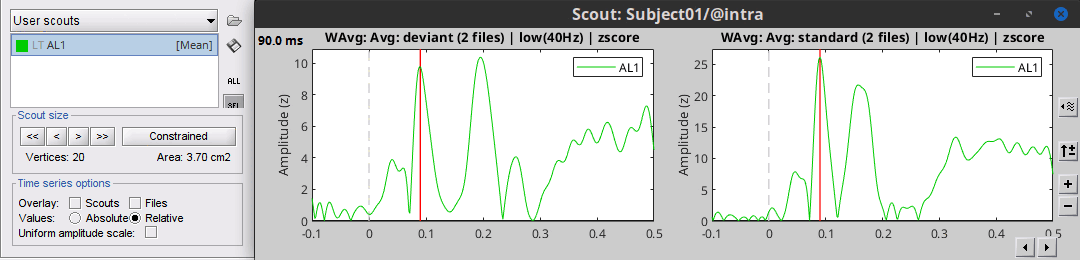
In the Record tab, you can use the button [=] to configure the amplitude of multiple axes.
Select it to force multiple graphs to use the same scale, unselect it to scale each graph separately.
You can find the same option in the figure popup menu: Figure > Uniform figure scale.
At the bottom of the Scout tab, select the option "Overlay: Files", then click [Display] again.
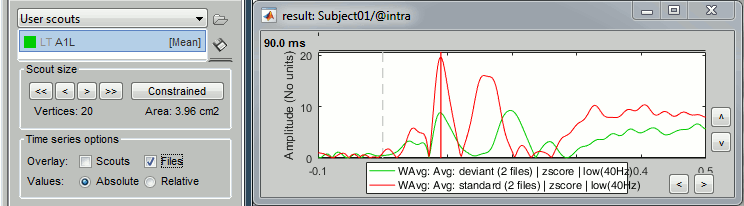
The Z-score value for the standard condition is a lot higher than the deviant condition because of the number of trials we used for computing the two averages (5x more trials for the standard). The SNR is higher with more trials, the baseline has less variance so the Z-score is higher.
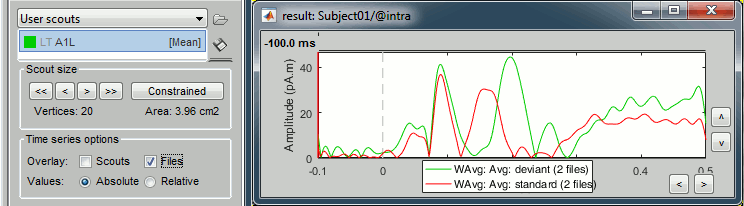
To overlay the results of two conditions averaged with very different numbers of trials, it makes more sense to display the scout time series for non-normalized maps (averaged current density).
Other regions of interest
- Let's place all the regions of interest starting from the easiest to identify.
Open the normalized average source files (standard and deviant), together with the average recordings for the deviant and standard condition for Run#01 for an easier time navigation.
In the Surface tab, smooth the cortical surface at 70%.
- For each region: go to the indicated time point, adjust the amplitude threshold in the Surface tab, identify the area of interest, click on its center, grow the scout, rename it.
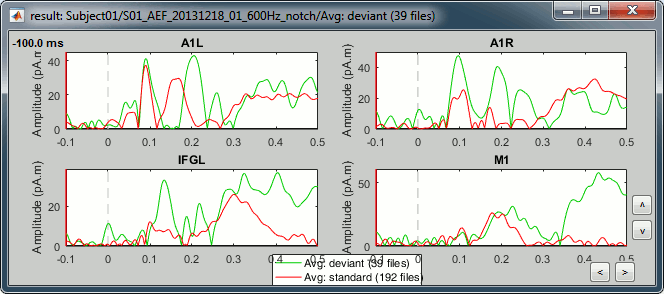
Grow all the regions to the same size: 20 vertices.
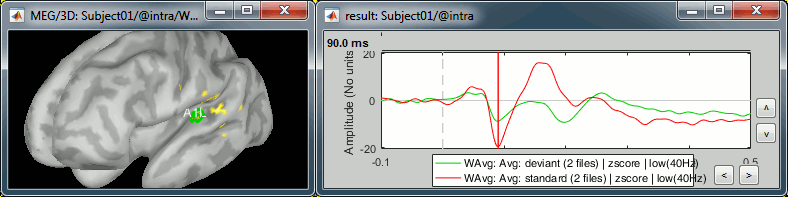
A1L: Left primary auditory cortex (Heschl gyrus) - Already marked.
- The most visible region in both conditions. Active during all the main steps of the auditory processing: P50, N100, MMN, P200, P300.
Standard condition, t=90ms, amplitude threshold=50%
A1R: Right primary auditory cortex (Heschl gyrus)
- The response in this region is not as obvious than A1L. These binaural auditory stimulations should be generating similar responses in both left and right auditory cortices at early latencies. Possible explanations for this observation:
- The earplug was not adjusted on the right side and the sound was not well delivered.
- The subject's hearing from the right ear is impaired.
- The response is actually stronger in the left auditory cortex for this subject.
- The orientation of the source makes it more difficult for the MEG sensors to capture.
Deviant condition, t=90ms, amplitude threshold=50%
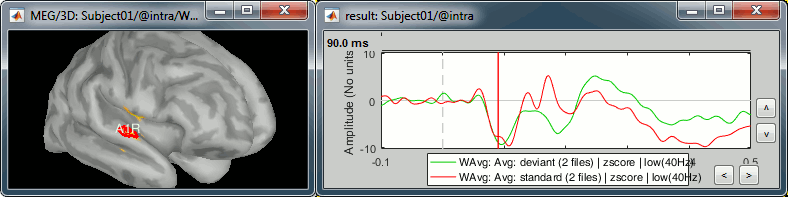
- The response in this region is not as obvious than A1L. These binaural auditory stimulations should be generating similar responses in both left and right auditory cortices at early latencies. Possible explanations for this observation:
IFGL: Left inferior frontal gyrus (Brodmann area 44)
- Involved in the auditory processing, particularly while processing irregularities.
- You can use the atlas "Brodmann-thresh" available in the Scout tab for identifying this region.
Deviant condition, t=130ms, amplitude threshold=30%
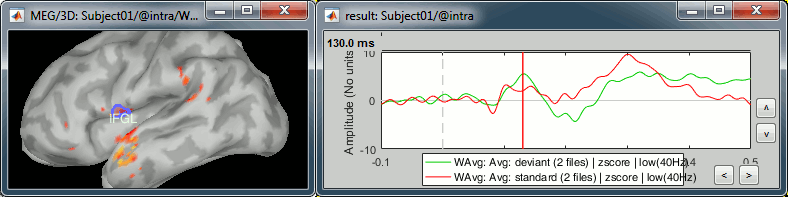
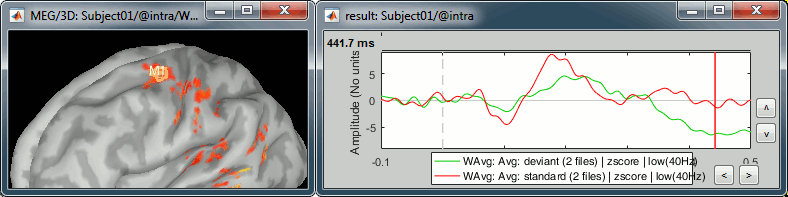
M1L: Left motor cortex
- The subject taps with the right index when a deviant is presented.
- The motor cortex responds at very early latencies together with the auditory cortex, in both conditions (50ms and 100ms). The subject is ready for a fast response to the task.
- At 170ms, the peak in the standard condition probably corresponds to an inhibition: the sound heard is not a deviant, there is no further motor processing required.
- At 230ms, the peak in the deviant condition is probably a motor preparation. At 350ms, the motor task begins, the subject moves the right hand (recorded reaction times 500ms +/- 108ms).
- We cannot expect to have clear responses during the motor response because of the averaging. The response times are variable, so in order to get a better representation of the regions involved we should import and average the trials based on the response triggers.
Deviant condition, t=440ms, amplitude threshold=25%
Multiple scouts
We can display the activity of multiple regions simultaneously.
- Close everything (toolbar button [X]).
Open the two normalized average source files (standard and deviant).
In the Scout tab, select the A1L and IFGL scouts simultaneously.
Select the option "Overlay:Scouts", do not select "Overlay:Files", Absolute values.
Click on [Display scouts time series].
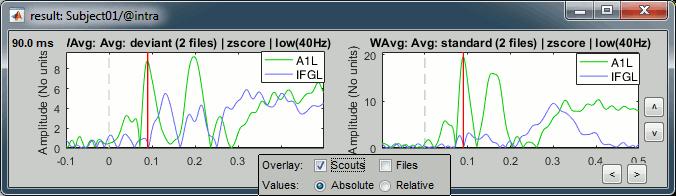
Now select the option "Overlay:Files", do not select "Overlay:Scouts", click on [Display].
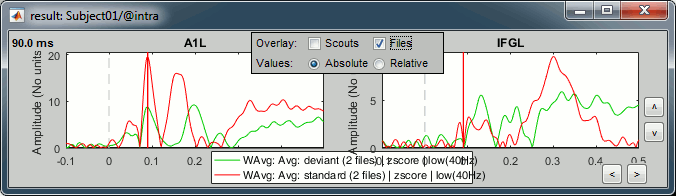
In both conditions, we observe similar early responses (P50 and N100), then it diverges.
In the deviant condition, we observe a pattern A1-IFG-A1 that is absent in the standard condition.
From the database explorer
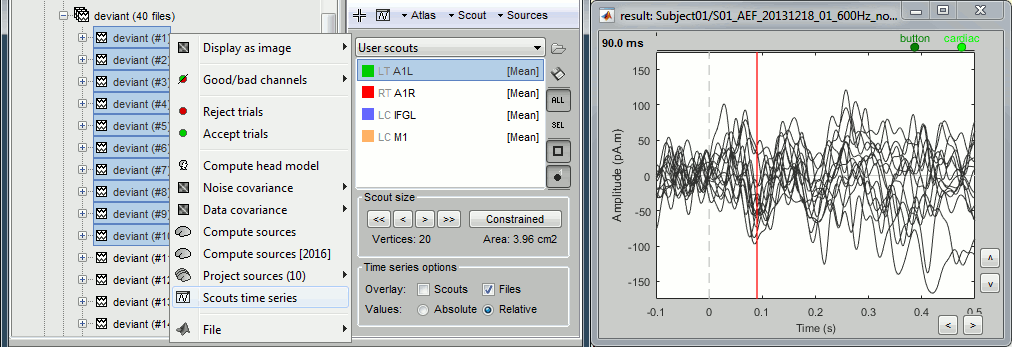
You have to display the sources on the cortex to create the scouts. But once they are created, you can directly display the scouts time series from the database explorer. It means that you can quickly compare the values for a scout between many different conditions without having to open them all.
- Close everything (toolbar button [X]).
Select the option "Overlay:Files", do not select "Overlay:Scouts".
Select the two average recordings in Run#01 folder > right-click > Scouts time series.
Note that this menu is present at all the levels in the database explorer.
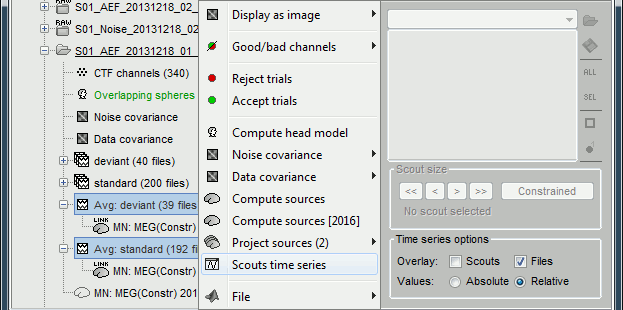
If no scout is currently loaded in the Scout tab, it shows all the scouts available in the surface.
If a surface is loaded, and at least one scout selected in the Scout tab, this popup menu would display only the selected scouts.
Once the list of scouts is loaded in the Scout tab, you can select one of them, and then display its values for all the trials of a single condition (overlay:files).
Sign flip
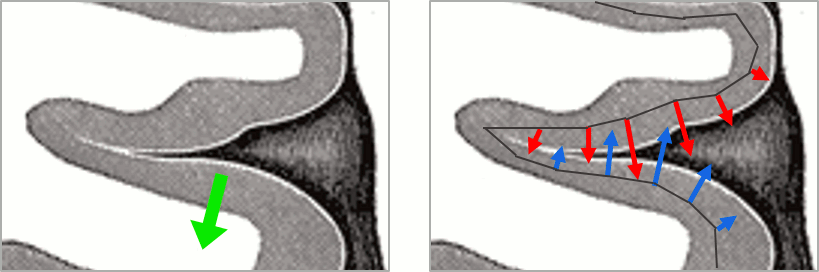
In the case of source models with constrained orientations (normal to the cortex), the sign of the current can be an issue. If the region is large enough to include vertices with normals in opposite directions, averaging the source values may cancel out the activity.
Let's use the example from the previous tutorial and consider that one scout contains all the dipoles corresponding to both the red arrows (positive values) and the blue arrows (negative values). If we average all the values from this scout, we get a value close to zero.
To avoid this, a mechanism was added in the scout calculation, to flip the sign of sources with opposite directions before the averaging. We start by finding the dominant orientation of the scout, then flip the sign of the values that are not in the same direction (scalar product of the orientations < 0).
If the sign of some sources is flipped, you get a message in the Matlab command window, for example:
BST> Flipped the sign of 7 sources.
Scout toolbar and menus
Menu: Atlas
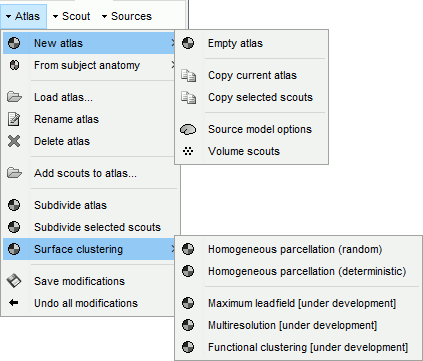
New atlas:
Empty atlas: Create a new empty atlas.
Copy atlas: Duplicate an atlas and all the scouts it contains.
Copy selected scouts: Create a new atlas and copy only the selected scouts to it.
Source model options: Create a special atlas "Source model", with which you can attribute different modeling constraints to each region, after merging the cortex with some subcortical regions. This is required to use the option "Custom head model" (mixed models).
Volume scouts: Create a volume atlas in order to define 3D scouts (volume head models).
From subject anatomy: Import as surface or volume scouts the ROIs defined in one of the volume parcellation available in the subject's anatomy (MNI-based atlases or subject parcellations from CAT12 or FreeSurfer).
Load atlas: Load ROIs or a cortical parcellation coming from FreeSurfer or BrainSuite as a new atlas.
Rename atlas: Change the name that appears in the atlas list to refer to the selected atlas.
Delete atlas: Delete the current atlas and all the scouts in contains.
Add scouts to atlas: Load ROIs or a cortical parcellation and add them to the current atlas.
Subdivide atlas: Splits all the scouts of the current atlas in smaller ROIs. Available options:
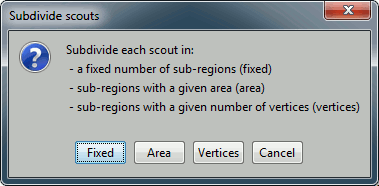
Subdivide selected scouts: Same thing, but processes only the selected scouts.
Surface clustering: Create a parcellation of the cortex surface and saves it as a new atlas.
Only the "Homogeneous parcellation" clustering is currently available.Save modifications: Force the current modifications to be saved to the cortex surface.
Undo all modifications: Restore the atlas the way it was the last time the surface was loaded.
Menu: Scout
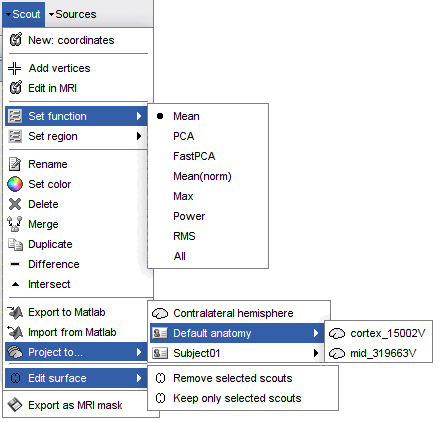
New: coordinates: Create a new scout starting from the vertex that is the closest to the specified coordinates. You can enter coordinates in MRI, SCS or MNI coordinates.
Add vertices: The user can select some points on the cortex surface and add them to the scout.
Equivalent: Click on Select point (first toolbar button) then select the scout in list.Edit in MRI: Open an interface to edit the selected scout as a 3D ROI, slice by slice. Only the vertices contained in the 3D mask are kept, all the volume information is lost. It means that the mask you intended to draw might be very different from what you get as a scout at the end. This is not a very reliable tool, as there is no direct correspondence between the volume and the surface.
Set function: Set the scout function for the selected scouts.
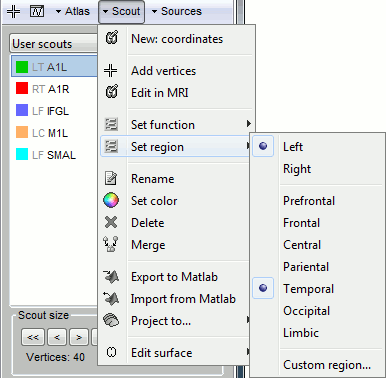
Set region: Set the cortical region for the selected scouts.
Rename: Rename the selected scout. Shortcut: double-click.
Set color: Change the display color of the selected scout.
Delete: Delete selected scouts. Shortcut: Delete key
Merge: Join two or more selected scouts.
Difference: With two scouts A and B selected, this menu removes the vertices included scouts A from scout B, or vice versa. The direction of the difference is asked in a dialog window.
Export to Matlab: Export the structures of the selected scouts to the Matlab environment and makes them accessible from the command window. This menu can be useful to get quickly the list of vertex indices or modify a scout manually.
Import from Matlab: Import scouts structures that you modified manually from your Matlab command window directly as new scouts.
Project to: Project the selected scout on another surface available in the database.
Project to: Contralateral hemisphere: Transfer scouts defined on one hemisphere to the other hemisphere. This feature is only available with FreeSurfer, and the recon-all segmentation pipeline must be executed with additional options. In the tutorial dataset and in the ICBM152 template anatomy, this option was not used, and therefore this menu generates an error. To get this contralateral projection to work, FreeSurfer recon-all must be executed again with the extra option, and the anatomy of the subject must be imported again. More information.
Edit surface: Create a new surface containing only the desired parts (remove or keep only the selected scouts). This is useful for instance for selecting one sub-cortical region from the Aseg FreeSurfer atlas (see the FreeSurfer tutorial).
Menu: Sources
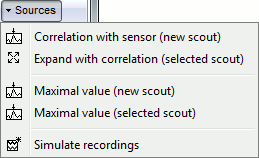
Correlation with sensor: Create a new scout with all the sources that are strongly correlated with a given sensor.
Expand with correlation: Computes the correlation between the values for the scout's seed (first point) and all the other sources. The sources that have correlation coefficient superior to a given threshold are added to the scout.
Maximal value (new scout): Find the vertex with the maximal intensity at the current time, and create a scout centered on it.
Maximal value (selected scout): Move the scout's seed to the source that has the maximum amplitude in the scout, at the current time.
Simulate recordings: Multiply the selected scouts with the forward model. Simulate the scalp data that would be recorded if only the selected cortex region was activated; all the other sources are set to zero. Create a new data file in the database.
If no scout selected: simulate recordings produced by the activity of the entire cortex.
Scout region
A scout is defined by its name, and it has several properties: a list of vertices and an aggregating function. These are usually enough to explore the activity at the cortex level the way we did it in these tutorials. An extra property can be defined on the scout: the explicit classification in a brain region. This property is used only in more advanced functional connectivity analysis, for the representation of the NxN connection graphs. It is introduced here for reference purpose.
A brain region in Brainstorm is following a hierarchy with three levels: hemisphere / lobe / sub-region. The definition at each level is optional: a region can be classified only at the hemisphere level, or at the hemisphere+lobe level, or none of them. It depends on level or hierarchy you are interested in to explore the connectivity graphs.
The region for a scout can be set with the Scout > Set region menus, and is encoded in a string that contains at least 2 characters: "HLxxx". H represents the hemisphere (L,R,U), L stands for the lobe (F,PF,C,P,T,O,L,U), and xxx for the sub-region name (optional). For both the hemisphere and the lobe, the value "U" stands for "Undefined", meaning that the classification is simply not set. The menu Set region>Custom region... lets you directly edit this string.
When set, the region string is shown before the scout name in the list, representing only the defined levels. It doesn't show the letters U for "undefined".
On the hard drive
The scouts are saved in the surface file on which they have been defined.
In the anatomy view, right-click on the selected cortex surface (cortex_15002V) > View file contents.
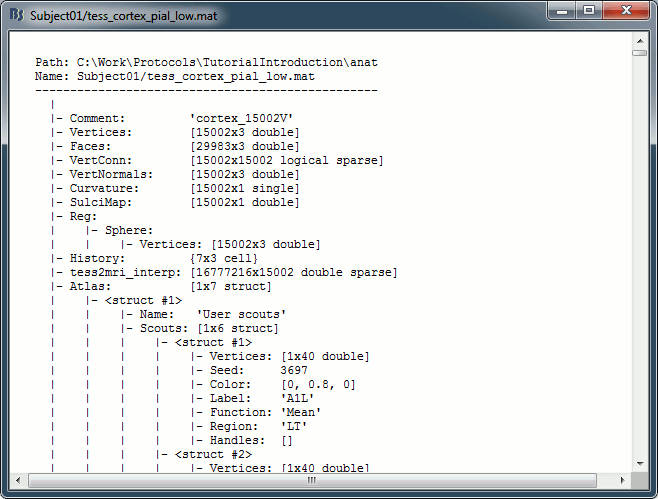
iAtlas: Index of the atlas that is currently selected for this surface.
Atlas: Array of structures, each entry is one menu in the drop-down list in the Scout tab.
Name: Label of the atlas (reserved names: "User scouts", "Structures", "Source model").
Scouts: Array of structures, one per scout in this atlas.
Vertices: Array of indices of the vertices that are part of the scout.
Seed: Index of the central point of the scout (or the most relevant one).
Color: [r,b,g] color array, with values between 0 and 1.
Label: Display name of the scout (must be unique in this atlas).
Function: Scout function {'Mean', 'PCA', 'FastPCA', 'Mean_norm', 'Max', 'Power', 'All'}
Region: Code name for indicating the anatomical region in which the scout is located.
Handles: Graphic handles if the scout is currently displayed (always empty in a file).
Useful functions
bst_scout_value: Combine multiple signals into one.
process_extract_scout: Process "Extract > Scouts time series"
view_scouts: Compute scouts time series and displays them in a new figure.
Additional documentation
Forum: Scout coordinates: http://neuroimage.usc.edu/forums/showthread.php?2375
Forum: Coordinates of the max value from a scout: https://neuroimage.usc.edu/forums/t/get-the-coordinates-of-a-max-value-from-scouts/4959/2
Forum: Importing other FreeSurfer atlases: https://neuroimage.usc.edu/forums/t/930