|
Size: 28506
Comment:
|
Size: 57586
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| <<HTML(<style>.backtick {font-size: 16px;}</style>)>><<HTML(<style>abbr {font-weight: bold;}</style>)>> <<HTML(<style>em strong {font-weight: bold; font-style: normal; padding: 2px; border-radius: 5px; background-color: #DDD; color: #111;}</style>)>> = MEG corticomuscular coherence = '''[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE] ''' ''Authors: Raymundo Cassani '' [[https://en.wikipedia.org/wiki/Corticomuscular_coherence|Corticomuscular coherence]] relates to the synchrony between electrophisiological signals (MEG, EEG or ECoG) recorded from the contralateral motor cortex, and EMG signal from a muscle during voluntary movement. This synchrony has its origin mainly in the descending communication in corticospinal pathways between primary motor cortex (M1) and muscles. This tutorial replicates the processing pipeline and analysis presented in the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|Analysis of corticomuscular coherence]] FieldTrip tutorial. <<TableOfContents(2,2)>> == Background == [[Tutorials/Connectivity#Coherence|Coherence]] is a classic method to measure the linear relationship between two signals in the frequency domain. Previous studies ([[https://dx.doi.org/10.1113/jphysiol.1995.sp021104|Conway et al., 1995]], [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Kilner et al., 2000]]) have used coherence to study the relationship between MEG signals from M1 and muscles, and they have shown synchronized activity in the 15–30 Hz range during maintained voluntary contractions. IMAGE OF EXPERIMENT, SIGNALS and COHERENCE |
<<HTML(<style>tt {font-size: 16px;}</style>)>><<HTML(<style>abbr {font-weight: bold;}</style>)>> <<HTML(<style>em strong {font-weight: normal; font-style: normal; padding: 2px; border-radius: 5px; background-color: #EEE; color: #111;}</style>)>> = Corticomuscular coherence (CTF MEG) = ''Authors: Raymundo Cassani, Francois Tadel, [[https://www.neurospeed-bailletlab.org/sylvain-baillet|Sylvain Baillet]].'' [[https://en.wikipedia.org/wiki/Corticomuscular_coherence|Corticomuscular coherence]] measures the degree of similarity between electrophysiological signals (MEG, EEG, ECoG sensor traces or source time series, especially over the contralateral motor cortex) and the EMG signals recorded from muscle activity during voluntary movements. Cortical-muscular signals similarities are conceived as due mainly to the descending communication along corticospinal pathways between primary motor cortex (M1) and the muscles attached to the moving limb(s). For consistency purposes, the present tutorial replicates, with Brainstorm tools, the processing pipeline "[[https://www.fieldtriptoolbox.org/tutorial/coherence/|Analysis of corticomuscular coherence]]" of the FieldTrip toolbox. <<TableOfContents(3,2)>> |
| Line 18: | Line 11: |
| The dataset is comprised of MEG (151-channel CTF MEG system) and bipolar EMG (from left and right extensor carpi radialis longus muscles) recordings from one subject during an experiment in which the subject had to lift her hand and exert a constant force against a lever. The force was monitored by strain gauges on the lever. The subject performed two blocks of 25 trials in which either the left or the right wrist was extended for about 10 seconds. In addition to the MEG and EMG signals, EOG signal was recorded to assist the removal of ocular artifacts. Only data for the left wrist will be analyzed in this tutorial. | The dataset is identical to that of the FieldTrip tutorial: [[https://www.fieldtriptoolbox.org/tutorial/coherence/|Analysis of corticomuscular coherence]]: * One participant, * MEG recordings: 151-channel CTF MEG system, * Bipolar EMG recordings: from left and right extensor carpi radialis longus muscles, * EOG recordings: used for detection and attenuation of ocular artifacts, * MRI: 1.5T Siemens system, * Task: The participant lifted their hand and exerted a constant force against a lever for about 10 seconds. The force was monitored by strain gauges on the lever. The participant performed two blocks of 25 trials using either their left or right hand. * Here we describe relevant Brainstorm tools via the analysis of the left-wrist trials. We encourage the reader to practice further by replicating the same pipeline using right-wrist trials! Corticomuscular coherence: * [[Tutorials/Connectivity#Coherence|Coherence]] measures the linear relationship between two signals in the frequency domain. * Previous studies ([[https://dx.doi.org/10.1113/jphysiol.1995.sp021104|Conway et al., 1995]], [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Kilner et al., 2000]]) have reported corticomuscular coherence effects in the 15–30 Hz range during maintained voluntary contractions. * TODO: IMAGE OF EXPERIMENT, SIGNALS and COHERENCE |
| Line 21: | Line 28: |
| * '''Requirements''': You should have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer. * '''Download the dataset''': * Download the `SubjectCMC.zip` file from FieldTrip FTP server: ftp://ftp.fieldtriptoolbox.org/pub/fieldtrip/tutorial/SubjectCMC.zip * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). * '''Brainstorm''': * Start Brainstorm (Matlab scripts or stand-alone version). * Select the menu '''File > Create new protocol'''. Name it '''TutorialCMC''' and select the options: '''No, use individual anatomy''', <<BR>> '''No, use one channel file per acquisition run'''. The next sections will describe how to link import the subject's anatomy, reviewing raw data, managing event markers, pre-processing, epoching, source estimation and computation of coherence in the sensor and sources domain. == Importing anatomy data == * Right-click on the '''TutorialCMC''' node then '''New subject > Subject01'''.<<BR>>Keep the default options you defined for the protocol. * Switch to the '''Anatomy''' view of the protocol. * Right-click on the '''Subject01''' node then '''Import MRI''': * Set the file format: '''All MRI file (subject space)''' |
'''Pre-requisites''' * Please make sure you have completed the [[Tutorials|get-started tutorials]], prior to going through the present tutorial. * Have a working copy of Brainstorm installed on your computer. * [[Tutorials/SegCAT12#Install_CAT12|Install the CAT12]] Brainstorm plugin, to perform MRI segmentation from the Brainstorm dashboard. '''Download the dataset''' * Download `SubjectCMC.zip` from the FieldTrip download page:<<BR>> https://download.fieldtriptoolbox.org/tutorial/SubjectCMC.zip * Unzip the downloaded archive file in a folder outside the Brainstorm database or app folders (for instance, directly on your desktop). '''Brainstorm''' * Launch Brainstorm (via the Matlab command line or the Matlab-free stand-alone version of Brainstorm). * Select from the menu '''File > Create new protocol'''. Name the new protocol `TutorialCMC` and select the options:<<BR>> No, use individual anatomy, <<BR>> No, use one channel file per acquisition run. == Importing anatomy == * Right-click on the newly created TutorialCMC node > '''New subject > Subject01'''.<<BR>>Keep the default options defined for the study (aka "protocol" in Brainstorm's vernacular). * Switch to the Anatomy view of the protocol (<<Icon(iconSubjectDB.gif)>>). * Right-click on the Subject01 > '''Import MRI''': * Select the file format: '''All MRI files (subject space)''' |
| Line 39: | Line 52: |
| * Compute MNI normalization, in the '''MRI viewer''' click on '''Click here to compute MNI normalization''', use the '''maff8''' method. When the normalization is complete, verify the correct location of the fiducials and click on '''Save'''. {{{#!wiki comment {{attachment:viewer_mni_norm.png}} |
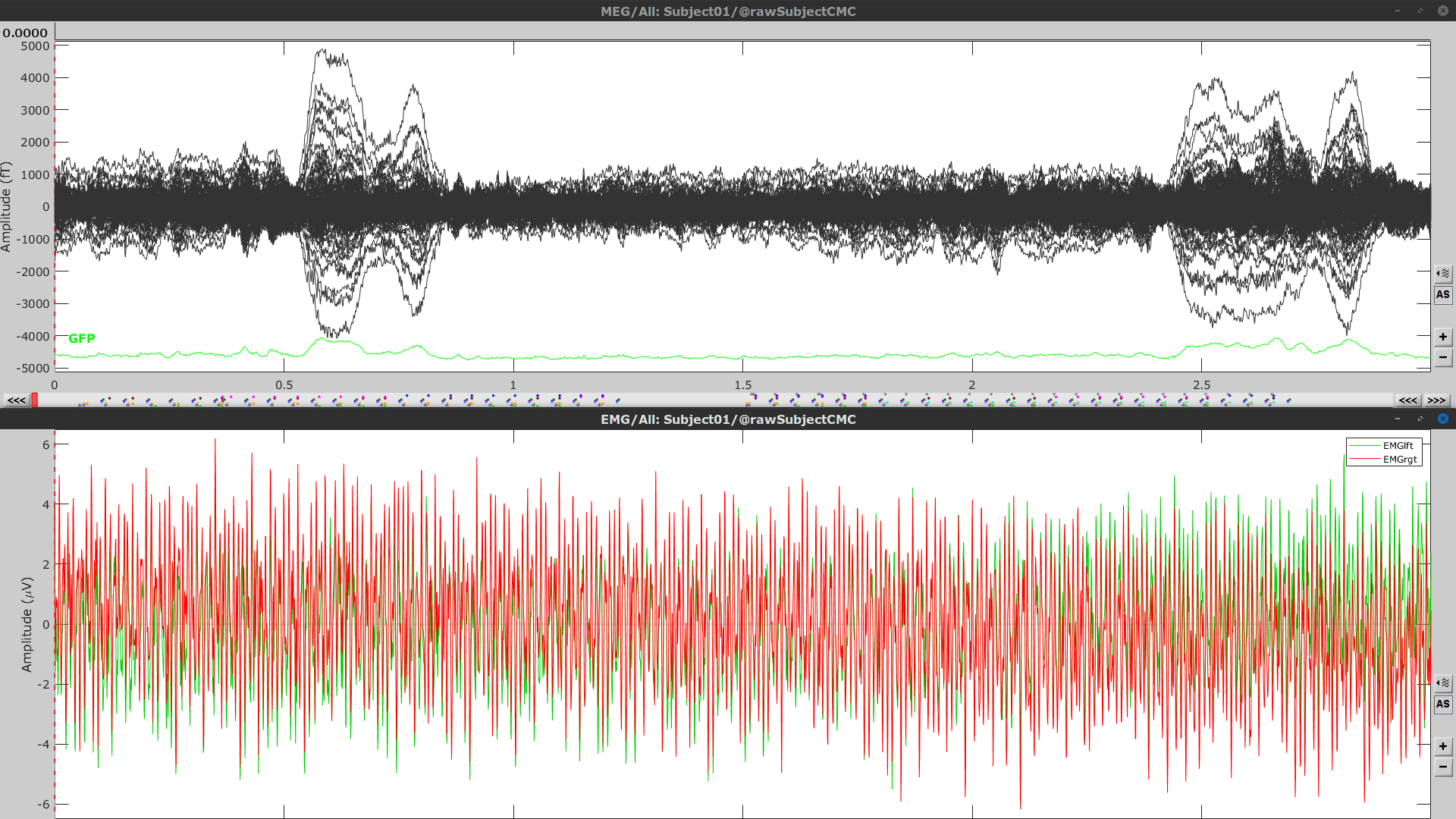
* This step will launch Brainstorm's MRI viewer, where coronal, sagittal and axial cross-sections of the MRI volume are displayed. note that [[CoordinateSystems|three anatomical fiducials]] (left and right pre-auricular points (LPA and RPA), and nasion (NAS)) are automatically identified. These fiducials are located near the left/right ears and just above the nose, respectively. Click on '''Save'''. <<BR>><<BR>> {{attachment:mri_viewer.gif||width="344",height="295"}} * In all typical Brainstorm workflows from this tutorial handbook, we recommend processing the MRI volume at this stage, before importing the functional (MEG/EEG) data. However, we will proceed differently, for consistency with the original FieldTrip pipeline, and readily obtain sensor-level coherence results. We will proceed with MRI segmentation below, before performing source-level analyses. * We still need to verify the proper geometric registration (alignment) of MRI with MEG. We will therefore now extract the scalp surface from the MRI volume. * Right-click on the MRI (<<Icon(iconMri.gif)>>) > MRI segmentation > '''Generate head surface'''. <<BR>><<BR>> {{attachment:head_process.gif}} * Double-click on the newly created surface to display the scalp in 3-D.<<BR>><<BR>> {{attachment:head_display.gif}} == MEG and EMG recordings == === Link the recordings === * Switch now to the '''Functional data '''view of your database contents (<<Icon(iconStudyDBSubj.gif)>>). * Right-click on Subject01 > '''Review raw file''': * Select the appropriate MEG file format: '''MEG/EEG: CTF(*.ds; *.meg4; *.res4)''' * Select the data file: `SubjectCMC.ds` * A new folder '''SubjectCMC '''is created in the Brainstorm database explorer. Note the "RAW" tag over the icon of the folder (<<Icon(iconRawFolderClose.gif)>>), indicating that the MEG files contain unprocessed, continuous data. This folder includes: * '''CTF channels (191)''' is a file with all channel information, including channel types (MEG, EMG, etc.), names, 3-D locations, etc. The total number of channels available (MEG, EMG, EOG etc.) is indicated between parentheses. * '''Link to raw file '''is a link to the original data file. Brainstorm reads all the relevant metadata from the original dataset and saves them into this symbolic node of the data tree (e.g., sampling rate, number of time samples, event markers). As seen elsewhere in the tutorial handbook, Brainstorm does not create copies by default of (potentially large) unprocessed data files ([[Tutorials/ChannelFile#Review_vs_Import|more information]]). . {{attachment:review_raw.png}} <<BR>> === MEG-MRI coregistration === * This registration step is to align the MEG coordinate system with the participant's anatomy from MRI ([[https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Automatic_registration|more info]]). Here we will use only the three anatomical landmarks stored in the MRI volume and specific at the moment of MEG data collection. From the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|FieldTrip tutorial]]:<<BR>>''"To measure the head position with respect to the sensors, three coils were placed at anatomical landmarks of the head (nasion, left and right ear canal). [...] During the MRI scan, ear molds containing small containers filled with vitamin E marked the same landmarks. This allows us, together with the anatomical landmarks, to align source estimates of the MEG with the MRI." '' * To visually appreciate the correctness of the registration, right-click on the '''CTF channels''' node > '''MRI registration > Check'''. This opens a 3-D figure showing the inner surface of the MEG helmet (in yellow), the head surface, the fiducial points and the axes of the [[CoordinateSystems#Subject_Coordinate_System_.28SCS_.2F_CTF.29|subject coordinate system (SCS)]].<<BR>><<BR>> {{attachment:fig_registration.gif||width="209",height="204"}} === Reviewing === * Right-click on the Link to raw file > '''Switch epoched/continuous''' to convert how the data i stored in the file to a continuous reviewing format, a technical detail proper to CTF file formatting. * Right-click on the Link to raw file > '''MEG > Display time series''' (or double-click on its icon). This will display data time series and enable the Time panel and the Record tab in the main Brainstorm window (see specific features in "[[Tutorials/ReviewRaw|explore data time series]]"). * Right-click on the Link to raw file > '''EMG > Display time series'''. . [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=timeseries_meg_emg.png|{{attachment:timeseries_meg_emg.png|https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=timeseries_meg_emg.png}}]] === Event markers === The colored dots at the top of the figure, above the data time series, indicate [[Tutorials/EventMarkers|event markers]] (or triggers) saved along with MEG data. The actual onsets of the left- and right-wrist trials are not displayed yet: they are saved in an auxiliary channel of the raw data named ''Stim''. To add these markers to the display, follow this procedure: * With the time series figure still open, go to the Record tab and select '''File > Read events from channel'''. Event channels = `Stim`, select Value, Reject events shorter than 12 samples. Click '''Run'''. . . {{attachment:read_evnt_ch.gif}} * The rejection of short events is nececessary in this dataset, because transitions between values in the Stim channel may span over several time samples. Otherwise, and for example, an event U1 would be created at 121.76s because the transition from the event U1025 back to zero features two unwanted values at the end of the event. The rejection criteria is set to 12 time samples (10ms), because the duration of all the relevant triggers is longer than 15ms. This value is proper to each dataset, so make sure you verify trigger detections from your own dataset.<<BR>><<BR>> {{attachment:triggers_min_duration.gif}} * New event markers are created and now shown in the Events section of the tab, along with previous event categories. In this tutorial, we will only use events '''U1''' through '''U25''', which correspond to the beginning of each of the 25 left-wrist trials of 10 seconds. Following FieldTrip's tutorial, let's reject trial #7, event '''U7'''. * Delete all unused events: Select all the events '''except''' '''U1-U6''' and '''U8-25''' (Ctrl+click / Shift+click), then menu '''Events > Delete group''' (or press the Delete key). * Merge events: Select all the event groups, then select from the menu '''Events > ''' Merge group > '''"Left"'''. A new event category called "Left" now indicate the onsets of 24 trials of left-wrist movements.<<BR>><<BR>> {{attachment:left_24.gif}} == Pre-processing == {{{#!wiki note In this tutorial, we will analyze only the '''Left''' trials (left-wrist extensions). In the following sections, we will process only the first '''330 s''' of the original recordings, where the left-wrist trials were performed. |
| Line 44: | Line 104: |
| . [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=viewer_mni_norm.png|{{attachment:viewer_mni_norm.png|https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=viewer_mni_norm.png}}]] * Once the MRI has been imported and normalized, we will segment the head and brain tissues to obtain the surfaces that are needed for a realistic [[Tutorials/TutBem|BEM forward model]]. * Right-click on the '''SubjectCMC''' MRI node, then '''MRI segmentation > FieldTrip: Tissues, BEM surfaces'''. * Select all the tissues ('''scalp''', '''skull''', '''csf''', '''gray''' and '''white'''). * Click '''OK'''. * For the option '''Generate surface meshes''' select '''No'''. * After the segmentation is complete, a '''tissues''' node will be shown in the tree. * Rick-click on the '''tissues''' node and select '''Generate triangular meshes''' * Select the 5 layers to mesh * Use the default parameters: '''number of vertices''': 10,000; '''erode factor''': 0; and '''fill holes factor''' 2. As output, we get a set of (head and brain) surface files that will be used for BEM computation. . {{attachment:import_result.png||width="40%"}} By displaying the surfaces, we can note that the '''cortex''', which is related to the gray matter (shown in red) overlaps heavily with the '''innerskull''' surface (shown in gray), so it cannot be used it for [[Tutorials/TutBem|BEM computation using OpenMEEG]]. However, as we are dealing with MEG signals, we can still compute the BEM with the [[Tutorials/HeadModel#Forward_model|overleaping-spheres method]], and obtain similar results. We can also notice that the '''cortex''' and '''white''' surfaces obtained with the method above do not register accurately the cortical surface, they can be used for [[Tutorials/TutVolSource|volume-based source estimation]], which is based on a volume grid of source points; but they do not be used for surface-based source estimation. Better surface surfaces can be obtained by doing MRI segmentation with [[Tutorials/SegCAT12|CAT12]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. . {{attachment:over_innerskul_cortex.png||width="50%"}} == Access the recordings == === Link the recordings === * Switch to the '''Functional data''' view (X button). * Right-click on the '''Subject01''' node then '''Review raw file''': * Select the file format: '''MEG/EEG: CTF(*.ds; *.meg4; *.res4)''' * Select the file: `SubjectCMC.ds` * A a new folder and its content is now visible in the database explorer: * The '''SubjectCMC''' folder represents the MEG dataset linked to the database. Note the tag "raw" in the icon of the folder, this means that the files are considered as new continuous files. * The '''CTF channels (191)''' node is the '''channel file''' and defines the types and names of channels that were recorded, the position of the sensors, the head shape and other various details. This information has been read from the MEG datasets and saved as a new file in the database. The total number of data channels recorded in the file is indicated between parenthesis (191). * The '''Link to raw file''' node is a '''link to the original file''' that was selected. All the relevant meta-data was read from the MEG dataset and copied inside the link itself (sampling rate, number of samples, event markers and other details about the acquisition session). As it is a link, no MEG recordings were copied to the database. When we open this file, the values are read directly from the original files in the .ds folder. [[Tutorials/ChannelFile#Review_vs_Import|More information]]. . {{attachment:review_raw.png}} <<BR>> === Display MEG helmet and sensors === * Right-click on the '''CTF channels (191)''' node, then '''Display sensors > CTF helmet''' and '''Display sensors > MEG ''' to show a surface that represents the inner surface the helmet, and the MEG sensors respectively. Try [[Tutorials/ChannelFile#Display_the_sensors|additional display menus]]. . {{attachment:helmet_sensors.png}} === Reviewing continuous recordings === * Right-click on the '''Link to raw file''' node, then '''Switch epoched/continuous''' to convert the file to '''continuous'''. * Right-click on the '''Link to raw file''' node, then '''MEG > Display time series''' (or double-click on the node). This will open a new time series figure and enable the '''Time panel''' and the '''Record''' tab in the main Brainstorm window. Controls in these two panels are used to [[Tutorials/ReviewRaw|explore the time series]]. * In addition we can display the EMG signals, right-click on the '''Link to raw file''' node, then '''EMG > Display time series'''. . [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=timeseries_meg_emg.png|{{attachment:timeseries_meg_emg.png|https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=timeseries_meg_emg.png}}]] === Event markers === The colored dots on top of the recordings in the time series figures represent the [[Tutorials/EventMarkers|event markers]] (or triggers) saved in this dataset. In addition to these events, the start of the either left or right trials is saved in the auxiliary channel named '''Stim'''. To add these markers: * With the time series figure open, in the '''Record''' tab go to '''File > Read events from channel'''. Now, in the options for the '''Read from channel''' process, set '''Event channels''': to `Stim`, select '''Value''', anc click '''Run'''. . {{attachment:read_evnt_ch.png}} New events will appear, from these, we are only interested in the events from '''U1''' to '''U25''' that correspond to the 25 left trials. Thus we will remove the other events, and merge the left trial events. * Delete all the other events: select the events to delete with '''Ctrl+click''', when done go the menu '''Events > Delete group''' and confirm. Alternatively, you can do '''Ctrl+A''' to select all the events and deselect the '''U1''' to '''U25''' events. * To be in line with the original FieldTrip tutorial, we will reject the trial 7. Select the events '''U1''' to '''U6''' and '''U8''' to '''U25''' events, then go the menu '''Events > Merge group''' and enter the label '''Left'''. . {{attachment:left_24.png}} These events are located at the beginning of the 10 s trials of left wrist movement. In the following steps we will compute the coherence for 1 s epochs for the first 8 s of the trial, thus we need extra events. * Duplicate 7 times the '''Left''' events by selecting '''Duplicate group''' in the '''Events''' menu. This will create the groups '''Left_02 to Left_08'''. * To each copy of the '''Left''' events, we will add a time off, 1 s for '''Left02''', 2 s for '''Left03''', and so on. Select the event group to add the offset, then go to the menu '''Events > Delete group'''. . {{attachment:dup_offset.png}} * Finally merge all the '''Left''' events into '''Left''', and select '''Save modifications''' in the '''File''' menu in the '''Record tab'''. . {{attachment:left_192.png}} === Keep relevant recordings === As only data for the left wrist will be analyzed, we will import only the first '''330 s''' of the original file and rewrite that segment as a binary continuous file, a raw file. This will help to optimize computation times and memory usage. * In the Process1 box: Drag and drop the '''Link to raw file''' node inside '''SubjectCMC'''. * Run process '''Import > Import recordings > Import MEG/EEG: Time''':<<BR>> * '''Subject name'''=`Subject01`, '''Condition name'''= `Left`, '''Time window'''=`0.0 - 330.0 s`, '''Split recordings'''=`0`, and check the three remaining options.<<BR>> . {{attachment:import330_process.png||width="50%"}} * Right-click on the '''Raw(0.00s,330.00s)''' node inside the newly created '''Left''' condition and select '''Review as raw'''. This will crate the condition '''block001''' with the link to the created raw file. . {{attachment:review_as_raw.png||width="50%"}} * To avoid any confusion later, delete the conditions '''SubjectCMC''' (which is a link to the original file), and the condition '''Left'''. Select both folders containing and press Delete (or right-click '''File > Delete'''). == Pre-process == |
|
| Line 130: | Line 105: |
| Let's start with locating the spectral components and impact of the power line noise in the MEG and EMG signals. * In the Process1 box: Drag and drop the '''Link to raw file''' node. |
* In the Process1 box: Drag and drop the '''Link to raw file'''. |
| Line 134: | Line 107: |
| * '''All file''', '''Window length='''`10 s`, '''Overlap'''=`50%`, and '''Sensor types'''=`MEG, EMG`<<BR>> * Double-click on the new PSD file to display it.<<BR>> |
* '''Time window''': `0-330 s` * '''Window length''':''' '''`10 s` * '''Overlap''': `50%` * '''Sensor types''': `MEG, EMG` . {{attachment:pro_psd.png||width="60%"}} * Double-click on the new PSD file to visualize the power spectrum density of the data.<<BR>> |
| Line 138: | Line 117: |
| * The PSD shows two groups of sensors, EMG on top and MEG in the bottom. Also, there are peaks at 50Hz and its harmonics. We will use notch filters to remove those frequency components from the signals. | * The PSD plot shows two groups of sensors: EMG (highlighted in red above) and the MEG spectra below (black lines). Peaks at 50Hz and harmonics (100, 150, 200Hz and above) indicate the European power line frequency and are clearly visible. We will now use notch filters to attenuate power line contaminants at 50, 100 and 150 Hz. * In the Process1 box: Drag and drop the '''Raw | clean''' node. |
| Line 141: | Line 121: |
| * '''Sensor types''' = `MEG, EMG` and '''Frequencies to remove (Hz)''' = `50, 100, 150, 200, 250, 300` . {{attachment:psd_process.png||width="50%"}} * Compute the PSD for the filtered signals to verify effect of the notch the filters. |
* Check '''Process the entire file at once''' * '''Sensor types''': `MEG, EMG` * '''Frequencies to remove (Hz)''': `50, 100, 150` . {{attachment:pro_notch.png||width="60%"}} * In case you get a memory error message:<<BR>>These MEG recordings have been saved before applying the CTF 3rd-order gradient compensation, for noise reduction. The compensation weights are therefore applied on the fly when Brainstorm reads data from the file. However, this requires reading all the channels at once. By default, the frequency filter are optimized to process channel data sequentially, which is incompatible with applying the CTF compensation on the fly. This setting can be overridden with the option '''Process the entire file at once''', which requires loading the entire file in memory at once, which may crash teh process depending on your computing resources (typically if your computer's RAM < 8GB). If this happens: run the process '''Artifacts > Apply SSP & CTF compensation''' on the file first, then rune the notch filter process without the option "Process the entire file at once" ([[https://neuroimage.usc.edu/brainstorm/Tutorials/TutMindNeuromag#Existing_SSP_and_pre-processing|more information]]). * A new folder named '''SubjectCMC_clean_notch''' is created. Obtain the PSD of these data to appreciate the effect of the notch filters. As above, please remember to indicate a '''Time window''' restricted from 0 to 330 s in the options of the PSD process.<<BR>><<BR>> |
| Line 147: | Line 131: |
| === Pre-process EMG === Two of the typical pre-processing steps for EMG consist in high-pass filtering and rectifying. * In the Process1 box: Drag and drop the '''Raw | notch(50Hz 100Hz 150Hz 200Hz 250Hz 300Hz)'''. |
=== EMG: Filter and rectify === Two typical pre-processing steps for EMG consist in high-pass filtering and rectifying. * In the Process1 box: drag and drop the '''Raw | notch(50Hz 100Hz 150Hz)''' node. |
| Line 152: | Line 136: |
| * '''Sensor types''' = `EMG`, '''Lower cutoff frequency''' = `10 Hz`, '''Upper cutoff frequency''' = `0 Hz` | * '''Sensor types''' = `EMG` * '''Lower cutoff frequency''' = `10 Hz` * '''Upper cutoff frequency''' = `0 Hz` |
| Line 155: | Line 142: |
* Run the pipeline |
|
| Line 157: | Line 147: |
| * Once the pipeline runs, the following report will appear. It contains '''warnings''' on the fact that applying absolute value to raw recordings is not recommended. We can safely ignore these warnings as we do want to rectify the EMG signals. . {{attachment:report.png||width="70%"}} * To do some cleaning we will remove the '''block001_notch_high''' condition. === Pre-process MEG === After applying the notch filter to the MEG signals, we still need to remove other artifacts, thus we will perform: 1. [[Tutorials/ArtifactsDetect|detection]] and [[Tutorials/ArtifactsSsp|removal of artifacts with SSP]] 1. [[Tutorials/BadSegments|detection of segments with other artifacts]]. ==== Detection and removal of ocular artifacts ==== * Display the MEG and EOG time series. Right-click on the filtered continuous file '''Raw | notch(...''' (in the '''block001_notch_high_abs''' condition), then '''MEG > Display time series''' and '''EOG > Display time series'''. * From the '''Artifacts''' menu in the '''Record''' tab, select '''Artifacts > Detect eye blinks''', and use the parameters: * '''Channel name'''= EOG, '''All file''' and '''Event name'''=blink. . {{attachment:detect_blink_process.png||width="50%"}} * As result, there will be 3 blink event groups. Review the traces of EOG channels and the blink events to be sure the detected events make sense. Note that the '''blink''' group contains the real blinks, and blink2 and blink3 contain mostly saccades. |
* Delete intermediate files that won't be needed anymore: Select folders '''SubjectCMC_notch''' and '''SubjectCMC_notch_high''',''' '''then press the Delete key (or right-click > File > Delete).<<BR>><<BR>> {{attachment:db_filters.gif}} === MEG: Blink SSP and bad segments === Stereotypical artifacts such eye blinks and heartbeats can be identified from their respective characteristic spatial distributions. Their contamination of MEG signals can then be attenuated specifically using Signal-Space Projections (SSPs). For more details, consult the specific tutorial sections about the [[Tutorials/ArtifactsDetect|detection]] and [[Tutorials/ArtifactsSsp|removal of artifacts with SSP]]. The present tutorial dataset features an EOG channel but no ECG. We will therefore only remove artifacts caused by eye blinks. ==== Blink correction with SSP ==== * Right-click on the pre-processed file > '''MEG > Display time series''' and '''EOG > Display time series'''. * In the Record tab: '''Artifacts > Detect eye blinks''', and use the parameters: * '''Channel name'''= `EOG` * '''Time window''' = `0 - 330 s` * '''Event name''' = `blink` . {{attachment:detect_blink_process.png||width="60%"}} * Three categories of blink events are created. Review the traces of the EOG channels around a few of these events to ascertain they are related to eye blinks. In the present case, we note that the '''blink''' group contains genuine eye blinks, and that groups blink2 and blink3 capture saccades. |
| Line 181: | Line 166: |
| * To [[Tutorials/ArtifactsSsp|remove blink artifacts with SSP]] go to '''Artifacts > SSP: Eyeblinks''', and use the parameters: * '''Event name'''=`blink`, '''Sensors'''=`MEG`, and check '''Compute using existing SSP''' . {{attachment:ssp_blink_process.png||width="50%"}} * Display the time series and topographies for the first two components. Only the first one is clearly related to blink artifacts. Select only component #1 for removal. |
* To [[Tutorials/ArtifactsSsp|remove blink artifacts with SSP]], go to '''Artifacts > SSP: Eye blinks''': * '''Event name'''=`blink` * '''Sensors'''=`MEG` . {{attachment:ssp_blink_process.png||width="60%"}} * Display the time series and topographies of the first two SSP components identified. In the present case, only the first SSP component can be clearly related to blinks: the percentage of variance explained is substantially higher than the other compoments', the spatial topography of the component is also typical of eye blinks, and the corresponding time series is similar to the EOG signal around blinks. Select only '''component #1''' for removal. |
| Line 190: | Line 176: |
| * Follow the same procedure for the other blink events. Note that none of first two components for the remaining blink events is clearly related to a ocular artifacts. This figure shows the first two components for the '''blink2''' group. . {{attachment:ssp_blink2.png||width="100%"}} . In this case, it is safer to unselect the '''blink2''' and '''blink3''' groups, rather than removing spatial components that we are not sure to identify. . {{attachment:ssp_active_projections.png||width="60%"}} * Close all the figures ==== Other artifacts ==== Here we will used automatic detection of artifacts, it aims to identify typical artifacts such as the ones related to eye movements, subject movement and muscle contractions. * Display the MEG and EOG time series. In the '''Record tab''', select '''Artifacts > Detect other artifacts''', use the following parameters: * '''All file''', '''Sensor types'''=`MEG`, '''Sensitivity'''=`3` and check both frequency bands '''1-7 Hz''' and '''40-240 Hz'''. . {{attachment:detect_other.png||width="50%"}} * While this process can help identify segments with artifacts in the signals, it is still advised to review the selected segments. After a quick browse, it can be noted that the selected segments correspond to irregularities in the MEG signal. * To label these events as bad, select them both in the events list and use the menu '''Events > Mark group as bad'''. Alternatively, you can rename the events and add the tag "bad_" in their name, it would have the same effect. |
* Close all visualization figures by clicking on the large '''×''' at the top-right of the main Brainstorm window. ==== Detection of "bad" data segments ==== Here we will use the [[Tutorials/BadSegments#Automatic_detection|automatic detection of artifacts]] to identify data segments contaminated by e.g., large eye and head movements, or muscle contractions. * Display the MEG and EOG time series. In the '''Record''' tab, select '''Artifacts > Detect other artifacts''' and enter the following parameters: * '''Time window''' = `0 - 330 s` * '''Sensor types'''=`MEG` * '''Sensitivity'''=`3` * Check both frequency bands '''1-7 Hz''' and '''40-240 Hz''' . {{attachment:detect_other.png||width="60%"}} * You are encouraged to review all the segments marked using this procedure. With the present data, all marked segments do contain clear artifacts. * Select the '''1-7Hz''' and '''40-240Hz''' event groups and select '''Events > Mark group as bad'''. Alternatively, you can add the prefix '''bad_''' to the event names. Brainstorm will automatically discard these data segments from further processing. |
| Line 214: | Line 195: |
| * Close all the figures, and save the modifications. == Importing the recordings == At this point we have finished with the pre-processing of our recordings. Many operations operations can only be applied to short segments of recordings that have been imported in the database. We will refer to these as "epochs" or "trials". Thus, the next step is to import the data taking into account the '''Left''' events. * Right-click on the filtered continuous file '''Raw | notch(...''' (in the '''block001_notch_high_abs''' condition), then '''Import in database'''. . {{attachment:import_menu.png||width="40%"}} * Set the import options as described below: . {{attachment:import_options.png||width="80%"}} The new folder '''block001_notch_high_abs''' appears in '''Subject01'''. It contains a copy of the channel file in the continuous file, and the '''Left''' trial group. By expanding the trial group, we can notice that there are trials marked with an interrogation sign in a red circle (ICON). These '''bad''' trials are the ones that were overleaped with the '''bad''' segments identified in the previous section. All the bad trials are automatically ignored in the '''Process''' panel. |
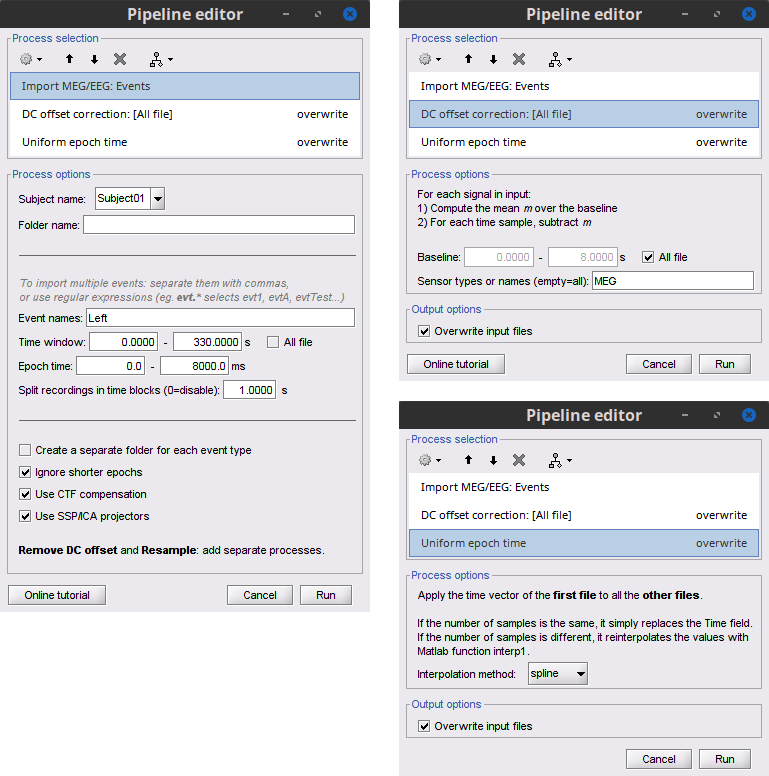
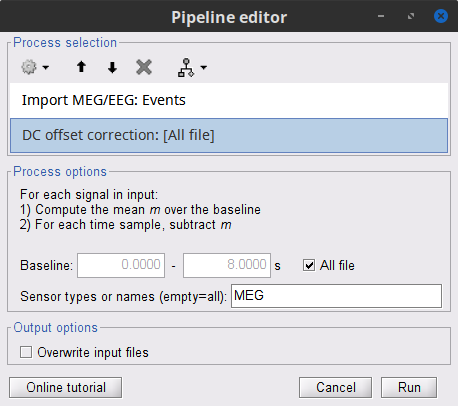
* Close all visualization windows and reply "Yes" to the save the modifications query. == Epoching == We are now finished with the pre-processing of EMG and MEG recordings. We will now extract and import specific data segments of interest into the Brainstorm database for further derivations. As mentioned previously, we will focus on the '''Left''' category of events (left-wrist movements). For consistency with the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|FieldTrip tutorial]], we will analyze 8s of recordings following each movement (from the original 10s around each trial), and split them in 1-s epochs. For each 1-s epoch, the DC offset will be also removed from each MEG channel. And finally, we will uniform the time vector for all 1-s epochs. This last steps is needed as the time vector of each 1-s vector is with respect to the event that was used to import the 8-s trial. * In the Process1 box: Drag-and-drop the pre-processed file. * Select the process '''Import > Import recordings > Import MEG/EEG: Events''': * '''Subject name''' = `Subject01` * '''Folder name''' = empty * '''Event names''' = `Left` * '''Time window''' = `0 - 330 s` * '''Epoch time''' = `0 - 8000 ms` * '''Split recordings in time blocks''' = `1 s` * Uncheck '''Create a separate folder for each event type''' * Check '''Ignore shorter epochs''' * Check '''Use CTF compensation''' * Check '''Use SSP/ICA projectors''' * Add the process '''Pre-process > Remove DC offset''': * '''Baseline''' = `All file` * '''Sensor types''' = `MEG` * Check '''Overwrite input files''' * Add the process '''Standardize > Uniform epoch time''': * '''Interpolation method''' = `spline` * Check '''Overwrite input files''' * Run the pipeline || {{attachment:pro_import.png}} || || {{attachment:pro_remove_dc.png}} || * A new folder '''SubjectCMC_notch_high_abs''' without the 'raw' indication is now created, which includes '''192 individual epochs''' (24 trials x 8 1-s epochs each). The epochs that overlap with a "bad" event are also marked as bad, as shown with an exclamation mark in a red circle (<<Icon(iconModifBad.gif)>>). These bad epochs will be automatically ignored by the '''Process1''' and '''Process2''' tabs, and from all further processing. |
| Line 231: | Line 233: |
| == Coherence (sensor level) == Here, we'll compute magnitude square coherence (MSC) between the '''left EMG''' and the signals from each of the MEG sensors. * In the '''Process1''' box, drag-and-drop the '''Left (192 files)''' trial group. Note that the number between square brackets is '''[185]''', as the 7 '''bad''' trials are ignored. |
==== Comparison with FieldTrip ==== The figures below show the EMG and MRC21 channels (a MEG sensor over the left motor-cortex) from the epoch #1.1, in Brainstorm (left), and from the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|FieldTrip tutorial]] (right). {{attachment:bst_ft_trial1.png||width="100%"}} == Coherence: EMG x MEG == Let's compute the '''magnitude-squared coherence (MSC)''' between the '''left EMG''' and the '''MEG''' channels. * In the Process1 box, drag and drop the '''Left (192 files)''' trial group. |
| Line 237: | Line 244: |
| * To compute the coherence between MEG and EMG signals. Run the process '''Connectivity > Coherence 1xN [2021]''' with the following parameters: * '''All file''', '''Source channel''' = `EMGlft`, do not '''Include bad channels''' nor '''Remove evoke response''', '''Magnitude squared coherence''', '''Window lenght''' = `0.5 s`, '''Overlap''' = `50%`, '''Highest frequency''' = `80 Hz`, and '''Average cross-spectra'''. * More details on the '''Coherence''' process can be found in the [[connectivity tutorial]]. . {{attachment:coh_meg_emgleft.png||width="40%"}} * Double-click on the resulting node '''mscohere(0.6Hz,555win): EMGlft''' to display the coherence spectra. Click on the maximum peak in the 15 to 20 Hz range, and press `Enter` to plot it in a new figure. The spectrum corresponds to channel MRC21, with its peak at 17.58 Hz. Right-click on the spectrum and select '''2D Sensor cap''' for a spatial visualization of the coherence results, alternatively, the short cut `Ctrl-T` can be used. |
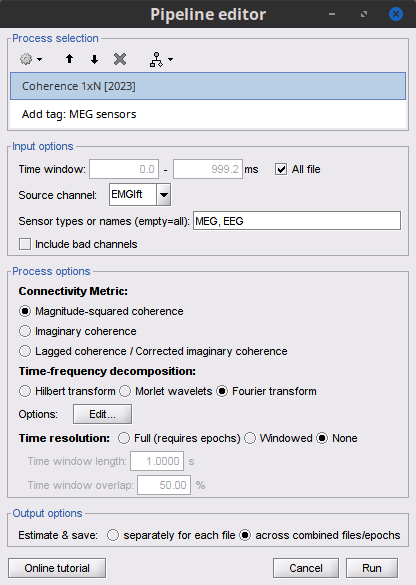
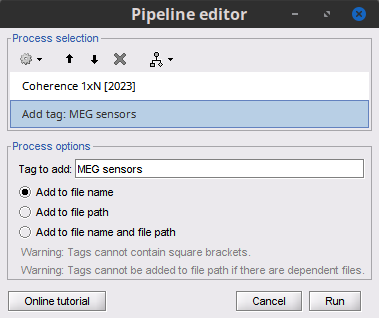
* Select the process '''Connectivity > Coherence 1xN [2023]''': * '''Time window''' = Check '''All file''' * '''Source channel''' = `EMGlft` * '''Sensor types''' = `MEG` * Do not check '''Include bad channels''' * Select '''Magnitude squared coherence''' as '''Connectivity metric''' * Select '''Fourier transform''' as '''Time-freq decomposition''' method * Click in the '''[Edit]''' button to set the parameters of the Fourier transform: * Select '''Matlab's FFT defaults''' as '''Frequency definition''' * '''FT window length''' = `0.5 s` * '''FT window overlap''' = `50%` * '''Highest frequency of interest''' = `80 Hz` * '''Time resolution''' = '''None''' * '''Estimate & save''' = '''across combined files/epochs''' {{{#!wiki note More details on the '''Coherence''' process can be found in the [[Tutorials/Connectivity#Coherence|Connectivity Tutorial]]. }}} * Add the process '''File > Add tag''' with the following parameters: * '''Tag to add''' = `MEG sensors` * Select '''Add to file name''' * Run the pipeline: || {{attachment:coh_meg_emgleft.png}} || || {{attachment:coh_meg_emgleft2.png}} || * Double-click on the resulting data node '''MSCoh-stft: EMGlft (185 files) | MEG sensors''' to display the MSC spectra. Click on the maximum peak in the 15 to 20 Hz range, and press `Enter` to display the spectrum from the selected sensor in a new window. This spectrum is that of channel '''MRC21''', and shows a prominent peak at 17.58 Hz. Use the frequency slider (under the Time panel) to explore the MSC output across frequencies. * Right-click on the spectrum and select '''2D Sensor cap''' for a topographical representation of the magnitude of the coherence results across the sensor array. You may also use the shortcut `Ctrl-T`. The sensor locations can be displayed with a right-click and by selecting '''Channels > Display sensors''' from the contextual menu (shortcut `Ctrl-E)`. |
| Line 247: | Line 277: |
| The results above are based in the identification of single peak, as alternative we can compute the average coherence in a given frequency band, and observe its topographical distribution. . {{attachment:pro_group_freq.png||width="40%"}} * In the '''Process1''' box, drag-and-drop the '''mscohere(0.6Hz,555win): EMGlft''', and add the process '''Frequency > Group in time or frequency bands''' with the parameters: * Select '''Group by frequency''', and type `cmc_band / 15, 20 / mean` in the text box. This will compute the mean of the coherence in the 15 to 20 Hz band. * Since our resulting file '''mscohere(0.6Hz,555win): EMGlft | tfbands''' is only one coherence value for each sensor, it is more useful to display in a spatial representation, for example '''2D Sensor cap'''. Sensor MRC21 is selected as reference. . {{attachment:res_coh_meg_emgleft1521.png||width="40%"}} In agreement with the literature, we observe a higher coherence between the EMG signal and the MEG signal from sensors over the contralateral primary motor cortex in the beta band range. In the next sections we will perform source estimation and compute coherence in the source level. == Source analysis == Before estimating the brain source, we need to compute the '''head model''' and the '''noise covariance'''. === Head model === The head model describes how neural electric currents produce magnetic fields and differences in electrical potentials at external sensors, given the different head tissues. This model is independent of sensor recordings. See the [[Tutorials/HeadModel|head model tutorial]] for more details. * In the '''block001_notch_high_abs''', right click the '''CTF channels (191)''' node and select '''Compute head model'''. Keep the default options: '''Source space''' = `Cortex`, and '''Forward model''' = `Overlapping spheres`. As result will have the '''Overlapping spheres''' head model in the database explorer. . {{attachment:pro_head_model.png||width="90%"}} |
* We can now average the magnitude of the MSC across the beta band (15-20 Hz). <<BR>>In the Process1 box, select the new '''mscohere''' file. * Run process '''Frequency > Group in time or frequency bands''': * Select '''Group by frequency bands''' * Type `cmc_band / 15, 20 / mean` in the text box. . {{attachment:pro_group_freq.png||width="60%"}} * The resulting '''MSCoh-stft:...|tfbands''' node contains one MSC value for each sensor (the MSC average in the 15-20 Hz band). Right-click on the file to display the 2D or 3D topography of the MSC beta-band measure. <<BR>><<BR>> {{attachment:res_coh_tfgroup.gif}} * Higher MSC values the EMG signal and MEG sensor signals map over the contralateral set of central sensors in the beta band. [[Tutorials/Connectivity#Sensor-level|Sensor-level connectivity]] can be ambiguous to interpret anaotmically though. We will now map the magnitude of EMG-coherence across the brain (MEG sources). == Source estimation == === MRI segmentation === We first need to extract the cortical surface from the T1 MRI volume we imported at the beginning of this tutorial. [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]] is a Brainstorm pluing that will perform this task in 30-60min. * Switch back to the Anatomy view of the protocol (<<Icon(iconSubjectDB.gif)>>). * Right-click on the MRI (<<Icon(iconMri.gif)>>) > '''MRI segmentation > CAT12''': * '''Number of vertices'''{{{: }}}`15000` * '''Anatomical parcellations''': `Yes` * '''Cortical maps''': {{{No}}} . {{attachment:cat12.png||width="100%"}} * Keep the low-resolution central surface selected as the default cortex ('''central_15002V'''). This surface is the primary output of CAT12, and is shown half-way between the pial envelope and the grey-white interface ([[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|more information]]). The head surface was recomputed during the process and duplicates the previous surface obtained above: you can either delete one of the head surfaces or ignore this point for now. * For quality control, double-click on the head and central_15002V surfaces to visualize them in 3D.<<BR>><<BR>> {{attachment:cat12_files.gif}} === Head models === We will perform source modeling using a [[Tutorials/HeadModel#Dipole_fitting_vs_distributed_models|distributed model]] approach for two different source spaces: the '''cortex surface''' and the entire '''MRI volume'''. Forward models are called ''head models'' in Brainstorm. They account for how neural electrical currents produce magnetic fields captured by sensors outside the head, considering head tissues electromagnetic properties and geometry, independently of actual empirical measurements ([[http://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel|more information]]). A distinct head model is required for the cortex surface and head volume source spaces. ==== Cortical surface ==== * Go back to the '''Functional data''' view of the database. * Right-click on the channel file of the imported epoch folder > '''Compute head model'''. * '''Comment''' = `Overlapping spheres (surface)` * '''Source space''' = `Cortex surface` * '''Forward model''' = `MEG Overlapping spheres`. . {{attachment:pro_head_model_srf.gif}} ==== Whole-head volume ==== * Right-click on the channel file again > '''Compute head model'''. * '''Comment''' = `Overlapping spheres (volume)` * '''Source space''' = `MRI volume` * '''Forward model''' = `Overlapping spheres`. * Select '''Regular grid''' and '''Brain''' * '''Grid resolution''' = `5 mm` . {{attachment:pro_head_model_vol.gif}} * The '''Overlapping spheres (volume)''' head model is now added to the database explorer. The green color of the name indicates this is the default head model for the current folder: you can decide to use another head model available by double clicking on its name. . {{attachment:tre_head_models.gif}} |
| Line 271: | Line 329: |
| For MEG it is recommended to derive the noise covariance from empty room recordings. However, as we don't have those recordings in the dataset, we compute the noise covariance from the MEG signals before the trials. See the [[Tutorials/NoiseCovariance|noise covariance tutorial]] for more details. * In the raw '''block001_notch_high_abs''', right click the '''Raw | notch(...''' node and select '''Noise covariance > Compute from recordings''''. As parameters select a '''Baseline''' from `12 - 30 s`, and the '''Block by block''' option. . {{attachment:pro_noise_cov.png||width="40%"}} * Lastly, we need to copy the '''Noise covariance''' node to the '''block001_notch_high_abs''' folder with the head model. This can be done with the shortcuts `Ctrl-C` and `Ctrl-V`. === Source estimation === With the help of the '''head model''' and the '''noise covariance''', we can solve the '''inverse problem''' by computing an '''inverse kernel''' that will estimate the brain activity that gives origin to the observed recordings in the sensors. See the [[Tutorials/SourceEstimation|source estimation tutorial]] for more details. * To compute the inversion kernel, right click in the '''Overlapping spheres''' head model and select '''Compute sources [2018]'''. With the parameters: '''Minimum norm imaging''', '''dSPM''' and '''Constrained'''. . {{attachment:pro_sources.png||width="40%"}} The inversion kernel '''dSPM-unscaled: MEG(Constr) 2018''' was created, and note that the each recordings node has an associated source link. . {{attachment:gui_inv_kernel.png||width="90%"}} == Coherence (source level) == {{{#!wiki warning '''[TO DISCUSS among authors]''' Better source localization can be obtained by performing MRI segmentation with CAT12. Although it adds between ~45min of additional processing. We may want to provide the already processed MRI. Thoughts? |
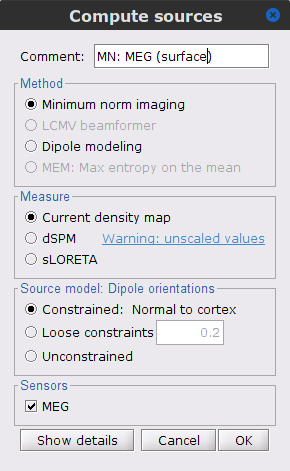
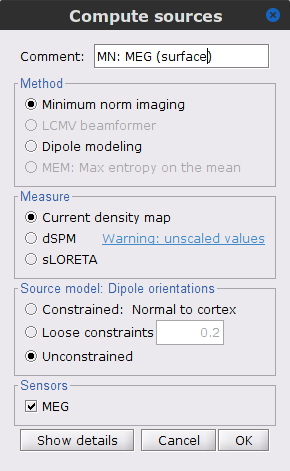
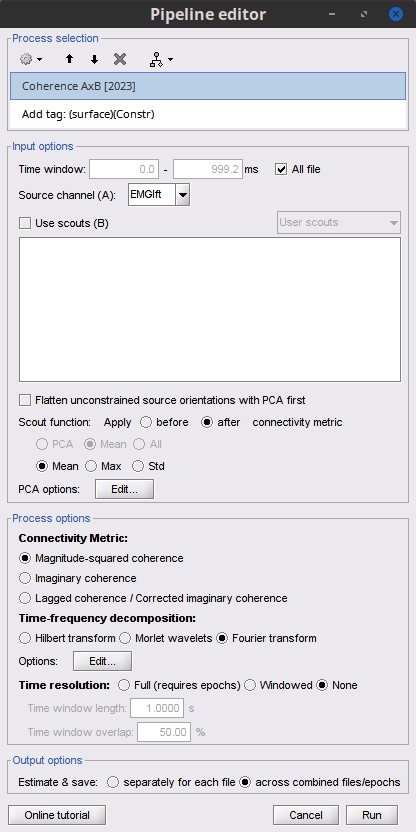
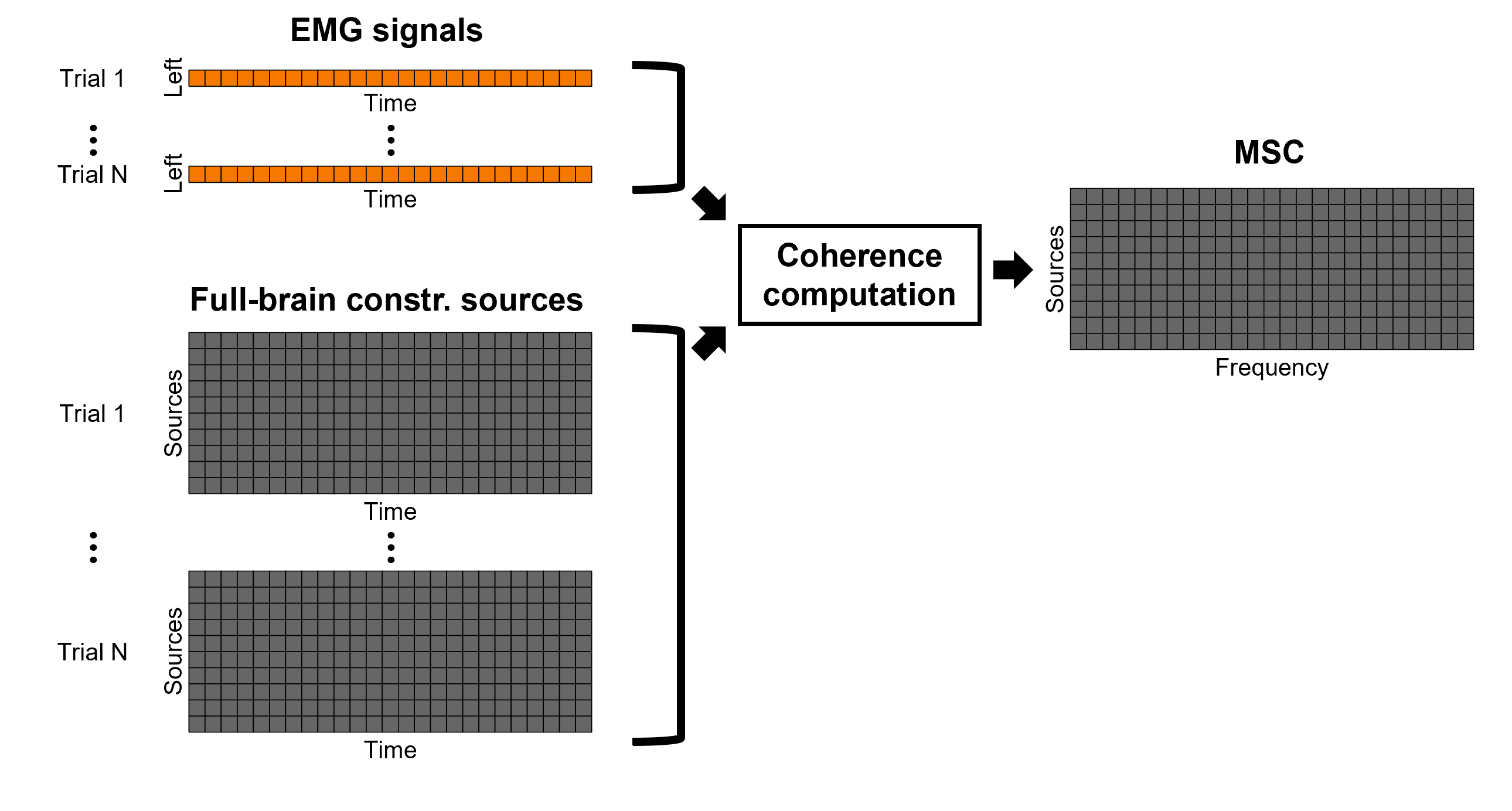
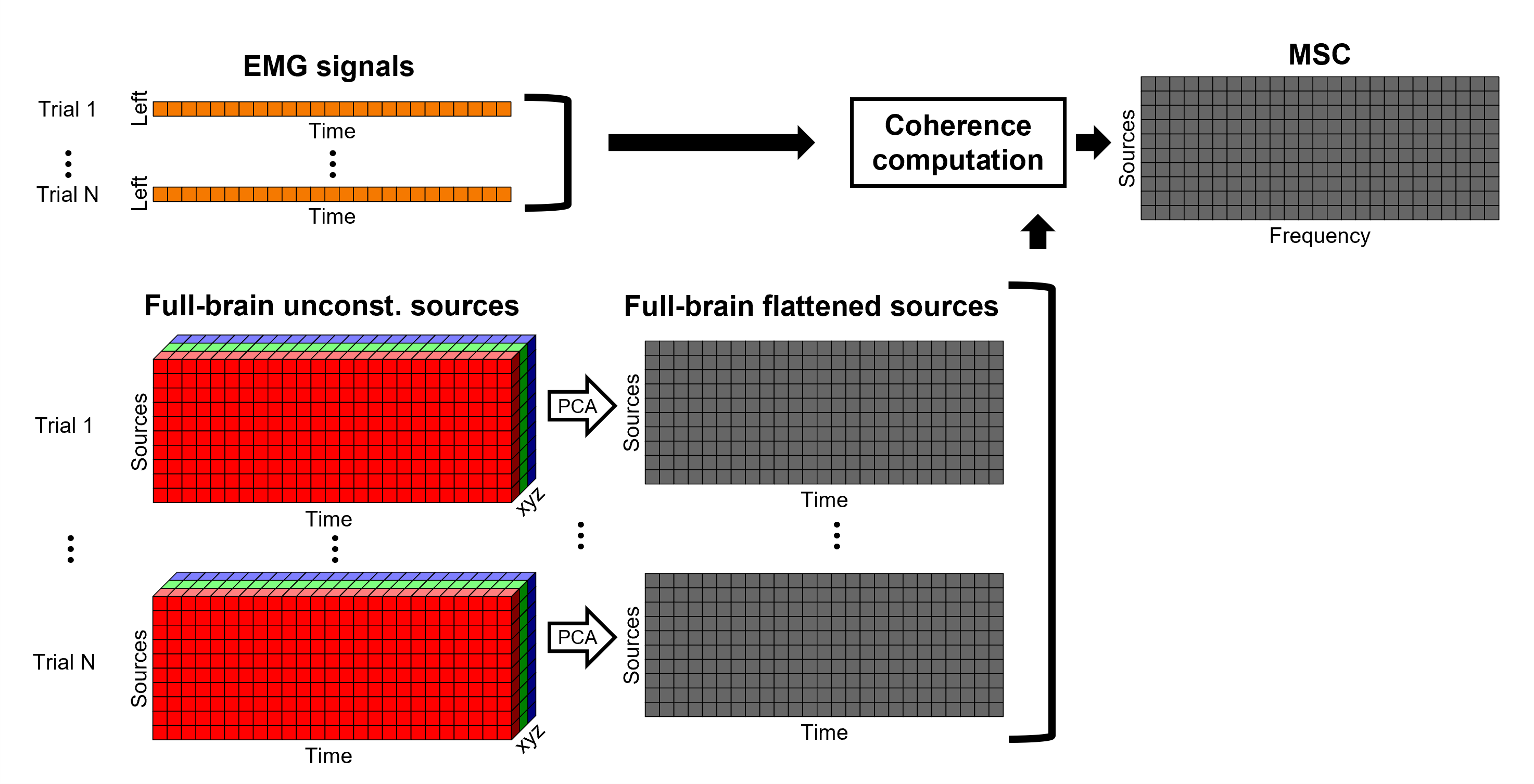
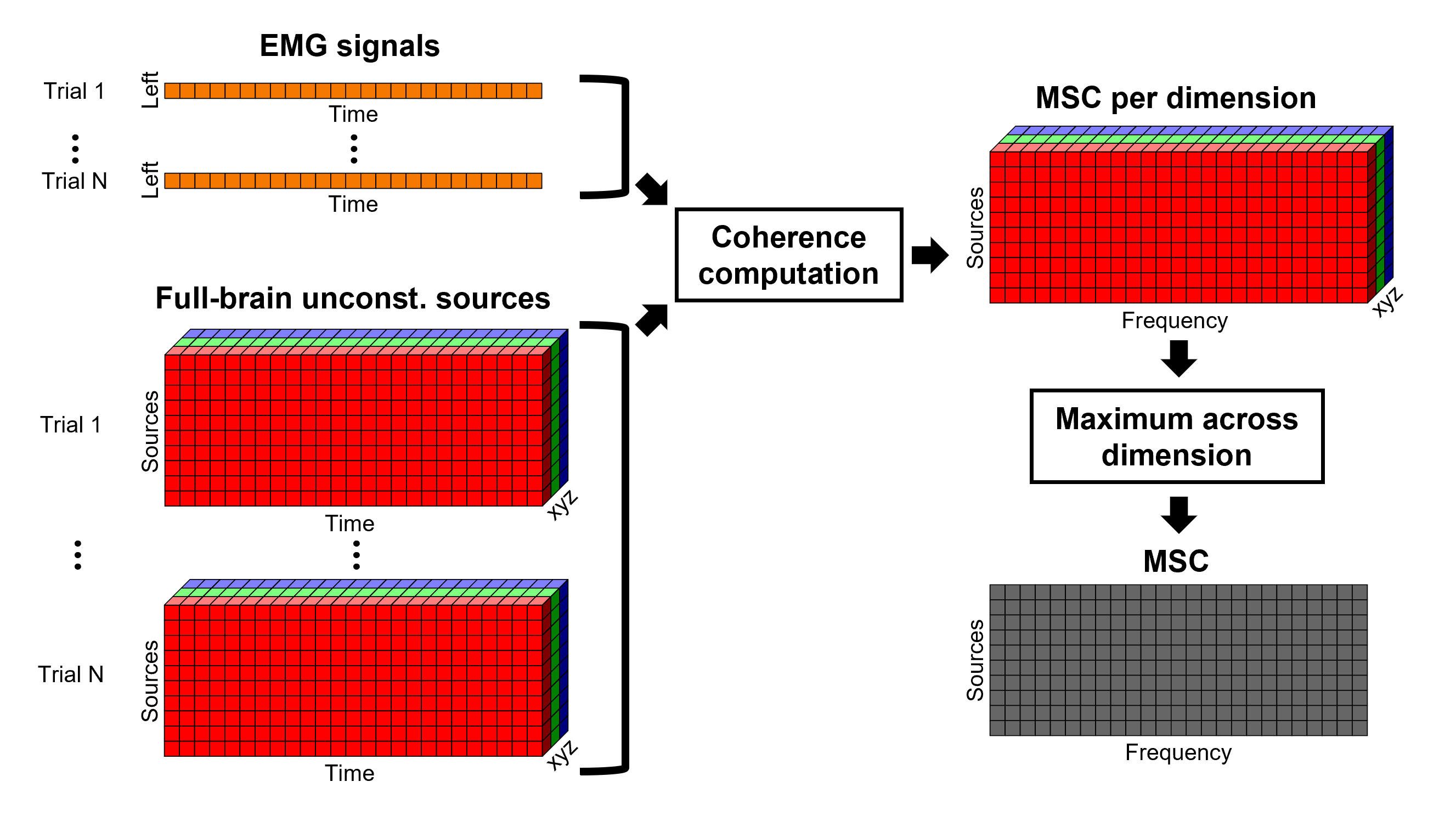
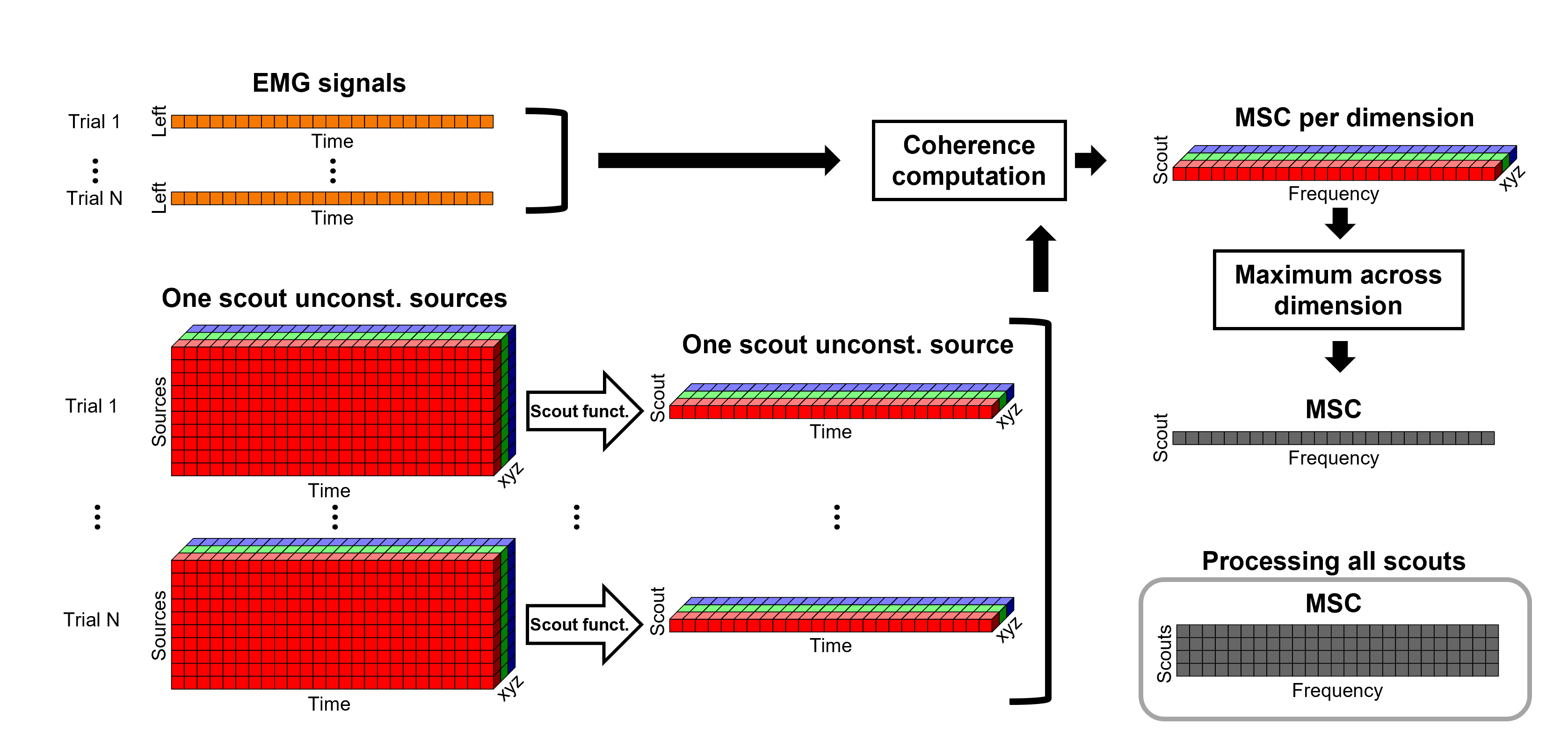
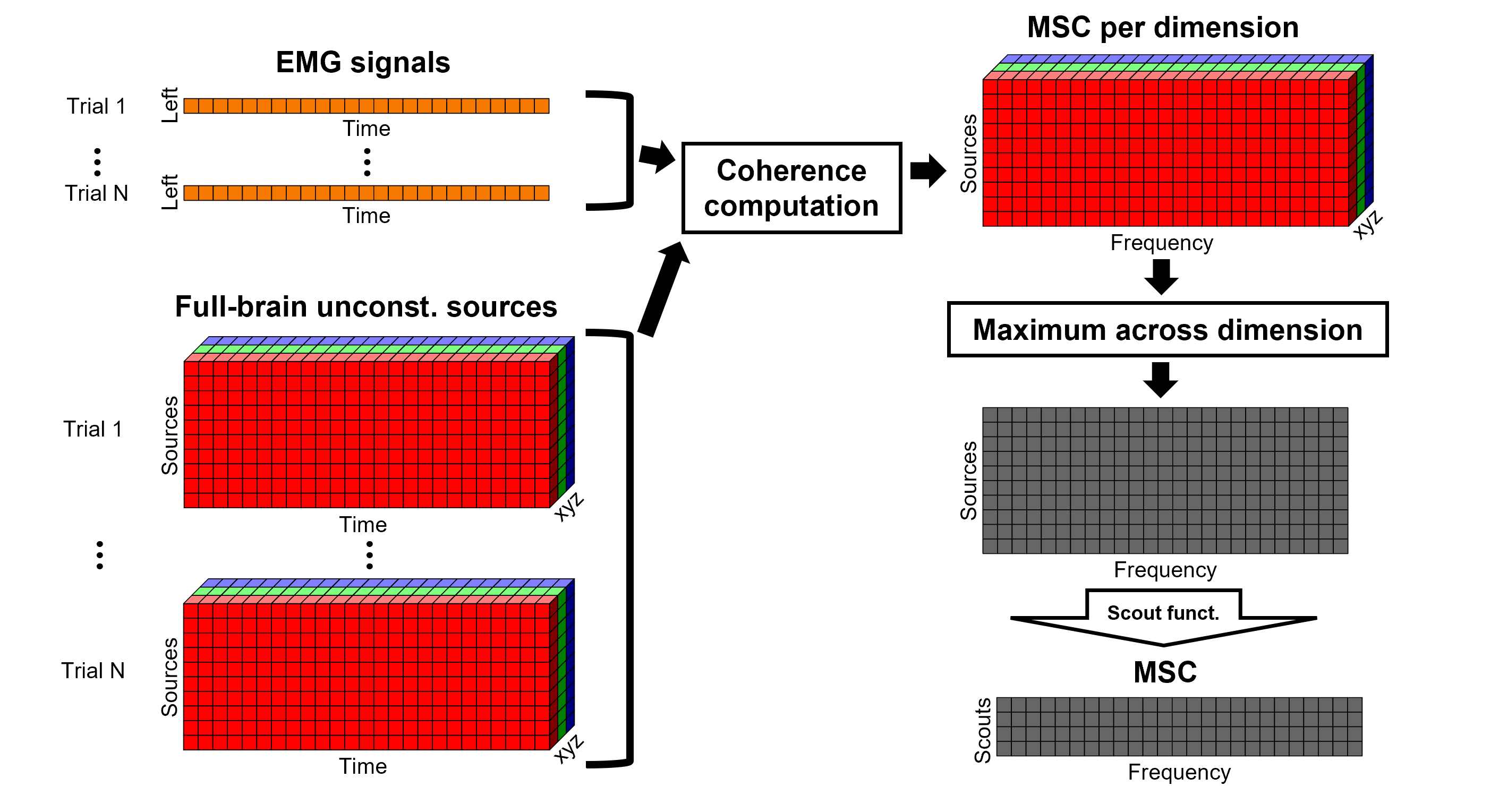
The [[Tutorials/NoiseCovariance#The_case_of_MEG|recommendation for MEG]] source imaging is to extract basic noise statistics from empty-room recordings. When not available, as here, resting-state data can be used as proxies for MEG noise covariance. We will use a segment of the MEG recordings, away from the task and major artifacts: '''18s-29s'''. * Right-click on the clean continuous file > '''Noise covariance > Compute from recordings'''.<<BR>><<BR>> {{attachment:pro_noise_cov.gif}} * Right-click on the Noise covariance (<<Icon(iconNoiseCov.gif)>>) > '''Copy to other folders'''.<<BR>><<BR>> {{attachment:tre_covmat.gif}} === Inverse models === We will now compute three inverse models, with different source spaces: cortex surface with '''constrained''' dipole orientations (normal to the cortex), cortex surface with '''unconstrained''' orientation, and MRI '''volume''' ([[Tutorials/SourceEstimation|more information]]). ==== Cortical surface ==== * Right-click on '''Overlapping spheres (surface)''' > '''Compute sources''': * '''Minimum norm imaging''' * '''Current density map''' * '''Constrained: Normal to the cortex''' * '''Comment''' = `MN: MEG (surface)` * Repeat the previous step, but this time select '''Unconstrained''' in the Dipole orientations field. || {{attachment:pro_sources_srfc.png}} || || {{attachment:pro_sources_srfu.png}} || ==== Whole-head volume ==== * Right-click on the '''Overlapping spheres (volume)''' > '''Compute sources:''' * '''Current density map''' * '''Unconstrained''' * '''Comment''' = `MN: MEG (volume)` . {{attachment:pro_sources_vol.png||width="40%"}} * Three imaging kernels (<<Icon(iconResultKernel.gif)>>) are now available in the database explorer. Note that each trial is associated with three source links (<<Icon(iconResultLink.gif)>>). The concept of imaging kernels is explained in Brainstorm's main tutorial on source mapping. . {{attachment:tre_sources.gif}} == Coherence: EMG x Sources == We can now compute the coherence between the EMG signal and all brain source time series, for each of the source models considered. Let's start with the '''cortical surface/constrained''' model. * To select the source maps to include in the coherence estimation, click on the [[Tutorials/PipelineEditor#Search_Database|Search Database]] button (<<Icon(iconZoom.gif)>>), and select '''New search'''. Set the parameters as shown below, and click on '''Search'''. . {{attachment:gui_search_srf.png||width="70%"}} * This creates a new tab in the database explorer, showing only the files that match the search criteria. . {{attachment:tre_search_srf.gif}} * Click the '''Process2''' tab at the bottom of the main Brainstorm window. * '''Files A''': Drag-and-drop the '''Left (192 files)''' group, select '''Process recordings''' (<<Icon(iconEegList.gif)>>). * '''Files B''': Drag-and-drop the '''Left (192 files)''' group, select '''Process sources''' (<<Icon(iconResultList.gif)>>). * Objective: Extract from the same files both the EMG recordings (Files A) and the sources time series (Files B), then compute the coherence measure between these two categories of time series. Note that the blue labels over the file lists indicate that there are 185 "good" files (7 bad epochs). . {{attachment:process2.png||width="80%"}} * Select the process '''Connectivity > Coherence AxB [2023]''': * '''Time window''' = '''All file''' * '''Source channel (A)''' = `EMGlft` * Uncheck '''Use scouts (B)''' * Check '''Flatten unconstrained source orientations''' * Click in the '''[Edit]''' button of the '''PCA options''': * Select '''across all epochs/files''' as PCA method * '''PCA time window''' = '''All file''' * Check '''Remove DC offset''' * '''Baseline''' = '''All file''' * Ignore the '''Scout function''' options as we are not using scouts * Select '''Magnitude squared coherence''' as '''Connectivity metric''' * Select '''Fourier transform''' as '''Time-freq decomposition''' method * Click in the '''[Edit]''' button to set the parameters of the Fourier transform: * Select '''Matlab's FFT defaults''' as '''Frequency definition''' * '''FT window length''' = `0.5 s` * '''FT window overlap''' = `50%` * '''Highest frequency of interest''' = `80 Hz` * '''Time resolution''' = '''None''' * '''Estimate & save''' = '''across combined files/epochs''' * Add the process '''File > Add tag''': * '''Tag to add''' = `(surface)(Constr)` * Select '''Add to file name''' * Run the pipeline || {{attachment:pro_coh_srf.png}} || || {{attachment:pro_coh_srf2.png}} || * Repeat the steps above to compute the EMG-sources coherence for the other source models: '''surface/unconstrained''' and '''volume''': <<BR>> * Edit the search criteria accordingly: Right-click on the search tab > '''Edit search'''. * This updates automatically the file selection in the Process2 tab. * Select the processes as indicated above and '''update''' the '''file tag''' according to the type of sources. {{{#!wiki caution The parameters '''Flatten unconstrained source orientations''' and '''PCA options''' did not have any impact on '''constrained''' sources, as there already consist on one time series per source location. However, when used with '''unconstrained''' sources, the three time series per source location are flattened into one before computing coherence. If the option '''Flatten unconstrained source orientations''' is unchecked for '''unconstrained''' sources, coherence is computed between the sensor and each of the three time series per source location, resulting in three coherence spectra which are collapsed into one using the '''maximum''' value for each frequency band. |
| Line 294: | Line 422: |
| From the earlier section [[#Importing_anatomy_data|importing anatomy data]], we can observe that the cortex surface has '''10,000''' vertices, thus as many sources were estimated. AS it can be seen, it is not practical to compute coherence between the left EMG signal and each source. A way to address this issue is with the use of regions of interest or [[Tutorials/Scouts|Scouts]]. * Double-clik the source link for any of the trials to visualize the source space. This will enable the tab '''Scout''' in the main Brainstorm window. . {{attachment:scout_tab.png||width="70%"}} * In the '''Scout''' tab select the menu '''Atlas > Surface clustering > Homogeneous parcellation (deterministic)''', and set the '''Number of scouts''' to `80`. . {{attachment:parcellation.png||width="70%"}} It is important to note that the coherence will be performed between a sensor signal (EMG) and source signals in the scouts. * Change to the '''Process2''' tab, and drag-and-drop the '''Left (192 files)''' trial group into the '''Files A''' and into the '''Files B''' boxes. And select '''Process recordings''' for Files A, and '''Process sources''' for Files B. |
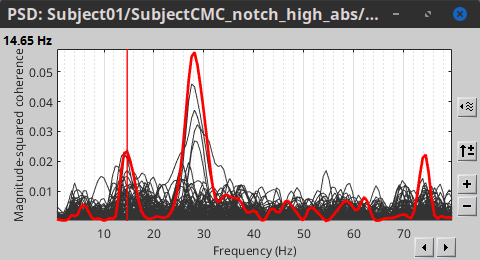
* Close the search tab. If the 3 new connectivity files <<Icon(iconConnect1.gif)>>) are not featured in the database explorer: refresh the dabase display by pressing '''[F5]''' or by clicking again on the selected button "Functional data". <<BR>><<BR>> {{attachment:tre_coh_src_files.gif}} === Surface === Double-click on the 1xN connectivity files for the two '''(surface)''' source spaces (constrained and unconstrained source orientations) to visualize the cortical maps. See main tutorial [[Tutorials/SourceEstimation#Display:_Cortex_surface|Display: Cortex surface]] for all available options. Pick the cortical location and frequency with the highest coherence value. * In the '''Surface''' tab: Smooth=30%, Amplitude=0%. * To compare visually different cortex maps, set manually the [[Tutorials/Colormaps|colormap]] range (e.g.`[0 - 0.07])` * Explore with coherence spectra with the '''frequency slider''' * The highest coherence value is located at '''14.65 Hz''', in the '''right primary motor cortex''' (precentral gyrus). To observe the coherence spectrum at a given location: right-click on the cortex > '''Source: Power spectrum'''. <<BR>><<BR>> {{attachment:res_coh_surf.gif}} * The constrained (top) and unconstrained (bottom) orientations coherence maps are qualitatively similar in terms of peak location and frequency. The unconstrained source map appears smoother because of the maximum aggregation across all three current directions at each brain location, as explained below. These maps results are similar to those obtained with the [[https://www.fieldtriptoolbox.org/tutorial/coherence/|FieldTrip tutorial]]. * To obtain the 3D coordinates of the peak coherence location: right-click on the figure > '''Get coordinates'''. Then click on the right motor cortex with the crosshair cursor. Let's keep note of these coordinates for later comparision with the whole-head volume results below. <<BR>><<BR>> {{attachment:res_get_coordinates.gif}} === Volume === * Double-click the 1xN connectivity file to display the '''(volume)''' source space. * Set the frequency to 14.65 Hz, and the data transparency to 20% (Surface tab). * Find the peak in the head volume by navigating in all 3 dimensions, or use the coordinates from the surface results as explained above. * Right-click on the figure > Anatomical atlas > None. The coherence value under the cursor is then shown on the top-right corner of the figure. <<BR>><<BR>> {{attachment:res_coh_vol.gif}} === Method for constrained sources === For cortical and ''' orientation-constrained''' sources, each vertex in the source grid is associated with '''1''' time series. As such, when coherence is computed with the EMG signal (also consisting of one time series), this result is '''1''' coherence spectrum per vertex. In other words, for each frequency bin, we obtain one coherence brain map. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_constr.png|{{attachment:diagram_1xn_coh_constr.png|width="100%"}}]] === Method for unconstrained sources === In the case of '''orientation-unconstrained''' (either surface or volume) sources, each vertex (source location) is associated with '''3''' time series, each one corresponding to the X, Y and Z directions. Thus, a '''dimension reduction across directions''' is needed. In this tutorial the dimension reduction consisted in first [[https://neuroimage.usc.edu/brainstorm/Tutorials/PCA#Unconstrained_source_flattening_with_PCA|flattening the 3D maps to a single orientation with the PCA method]]. This results in a source model similar to that with orientation-constrained sources, thus, coherence is then computed as with ''' orientation-constrained''' (described above). '''PCA Options''': If PCA is selected as the scout function or for flattening unconstrained maps, [[https://neuroimage.usc.edu/brainstorm/Tutorials/PCA#PCA_options|additional options]] are available through this ''Edit'' button. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_flattened.png|{{attachment:diagram_1xn_coh_flattened.png||width="100%"}}]] {{{#!wiki caution Another alternative to address the issue of the dimension reduction across directions when estimating coherence with '''unconstrained sources''' is to compute coherence between the EMG signal (one time series) and each of the '''3''' time series (one for each source orientation: x, y and z) associated with each source location, resulting in '''3''' coherence spectra per source location. Then, these coherence spectra are collapsed into a single coherence spectrum by keeping the '''maximum''' coherence value across those in the three directions for each frequency bin. The choice of the maximum statistic is empirical: it is not rotation invariant (if the specifications of the NAS/LPA/RPA fiducials change, the x,y,z axes also change, which may influence connectivity measures quantitatively). Nevertheless, our empirical tests showed this option produced smooth and robust (to noise) connectivity estimates. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_unconstr.png|{{attachment:diagram_1xn_coh_unconstr.png||width="100%"}}]] Below the comparison of the coherence between the EMG sensor and '''unconstrained surface''' sources using: '''PCA flattening in the source space''' (left) and '''"Flattening of the coherence, by maximum coherence across directions" ''' [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_unconstr.png|{{attachment:diagram_1xn_coh_unconstr.png||width="100%"}}]] }}} == Coherence: EMG x Scouts == We have now obtained coherence spectra at each of the 15002 brain source locations. This is a very large amount of data to shift through. We therefore recommend to further reduce the dimensionality of the source space by using a [[Tutorials/Scouts#Scout_toolbar_and_menus|surface-]] or [[Tutorials/DefaultAnatomy#MNI_parcellations|volume-]]parcellation scheme. In Brainstorm vernacular, this can be achieved via an '''atlas''' consisting of '''scout regions'''. See the [[Tutorials/Scouts|Scout Tutorial]] for detailed information about atlases and scouts in Brainstorm. To compute scout-wise coherence, it is necessary to specify two parameters to indicate how the data is aggregated in each scout: 1. The [[https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#Scout_function|scout function]] (for instance, the mean), and 2. When the scout aggregation procedure if performed, either '''before''' or '''after''' the coherence computation). === Method for constrained sources === When the '''scout function''' is used with '''constrained''' sources, the computation of coherence is carried as follows: * If the scout function is applied '''before''': The scout function (e.g., mean) is applied for all the source time series in the scout; resulting in '''1''' time series per scout. This scout time series is then used to compute coherence with the reference signal (here, with EMG) resulting in '''1''' coherence spectrum per scout. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_bef.png|{{attachment:diagram_1xn_coh_sct_bef.png||width="100%"}}]] * If the scout function applied '''after''': coherence is first computed between the reference signal (here, EMG) and each source time series in the scout, resulting in '''N''' coherence spectra, with '''N''' being the number of sources in the scout. Finally the scout function is applied to the resulting coherence spectra for each source within the scout to obtain one coherence spectrum per scout. This is a considerably more greedy option that the '''"before"''' option above, as it computes coherence measures between the EMG signal and each of the 15000 elementary source signals. This requires more computing resources; see [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#Source-level|introduction]] for an example. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_aft.png|{{attachment:diagram_1xn_coh_sct_aft.png||width="100%"}}]] === Method for unconstrained sources === As [[XX|described above]], when computing coherence with '''unconstrained sources''' it is necessary to perform dimension reduction across directions. When this reduction is done by first '''flattening''' the unconstrained sources, is performed on '''unconstrained sources''' coherence computations is carried out as described in the [[XX|previous section]]. However, if the sources are not flattened before computing coherence is carried out as follows: * If the scout function is applied '''before''', the scout function (e.g., mean) is applied for each direction on the elementary source time series in the scout; resulting in '''3''' time series per scout, one time series per orientation (x, y, z). These scout time series are then used to compute coherence with the reference signal (here, with EMG), and the coherence spectra for each scout are aggregated across the x, y and z dimensions, with the '''max''' function [[#Method|as shown previously]], to obtain '''1''' coherence spectrum per scout. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_bef.png|{{attachment:diagram_1xn_coh_sct_bef.png||width="100%"}}]] * If scout function is applied '''after''', coherence is computed between the reference signal (here, EMG) and each elementary sources (number of sources in each scout x 3 orientations). Then the coherence spectra for each elementary source are aggregated across the x, y and z dimensions. Finally the scout function is applied to the resulting coherence spectra for each elementary source within the scout. This is a considerably more greedy option that the "before" option above, as it computes coherence measures between the EMG signal and each of the 45000 elementary source signals. This requires more computing resources; see [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#Source-level|introduction]] for an example. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_aft.png|{{attachment:diagram_1xn_coh_sct_aft.png||width="100%"}}]] === Example === Here, we will compute the coherence from scouts, using '''mean''' as scout function with the '''before''' option. We will use the [[https://www.biorxiv.org/content/biorxiv/early/2017/07/16/135632.full.pdf|Schaefer 100 parcellation]] atlas on to the orientation-constrained source map. * Use Search Database (<<Icon(iconZoom.gif)>>) to select the '''Left''' trials with their respective '''(surface)(Constr)''' source maps, as shown in the [[#Coherence_1xN_.28source_level.29|previous section]]. * In the Process2: '''Left '''trial group into both the '''Files A''' and '''Files B''' boxes. Select '''Process recordings''' (<<Icon(iconEegList.gif)>>) for Files A, and '''Process sources''' (<<Icon(iconResultList.gif)>>) for Files B. |
| Line 310: | Line 505: |
| * Run the process '''Connectivity > Coherence AxB [2021]''' with the following parameters: * '''All file''', '''Source channel (A)''' = `EMGlft`, check '''Use scouts (B)''' * Select `Surface clustering: 80` in the drop-down list, and select all the scouts (shortcut `Ctrl-A`). * '''Scout function''' = `Mean`, and '''When to apply''' = `Before`. Do not '''Remove evoked responses from each trial'''. * '''Magnitude squared coherence''', '''Window length''' = `0.5 s`, '''Overlap''' = `50%`, '''Highest frequency''' = `80 Hz`, and '''Average cross-spectra'''. . {{attachment:pro_coh_ab.png||width="40%"}} === Results with FieldTrip MRI segmentation === * Double-click on the resulting node '''mscohere(0.6Hz,555win): Left (#1)''' to display the coherence spectra. Also open the result node as image with '''Display image''' in its context menu. * To verify the location of the scouts on the cortex surface, double-click the source link for any of the trials. In the '''Surface''' tab, set the '''Amplitude''' threshold to `100%` to hide all the cortical activations. Lasly, in the '''Scouts''' tab, select the '''Show only the selected scouts''' and the '''Show/hide the scout labels'''. Note that the plots are linked by the scout selected in the '''image''' representation of the coherence results. This will enable the tab '''Scout''' in the main Brainstorm window. === Results with CAT12 MRI segmentation === == Script == This should be label as advanced. |
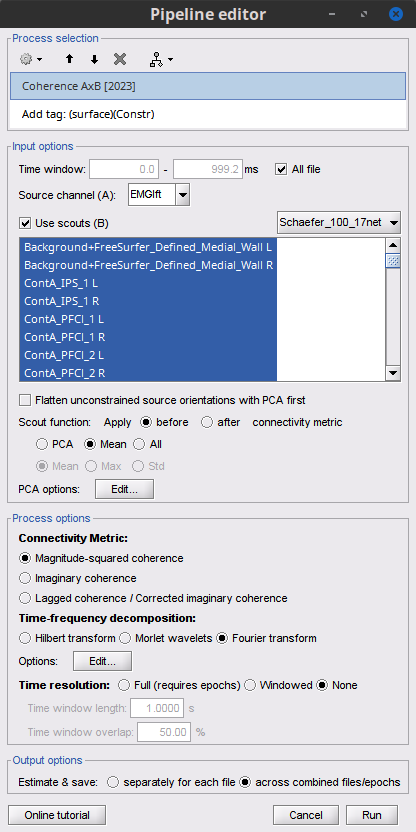
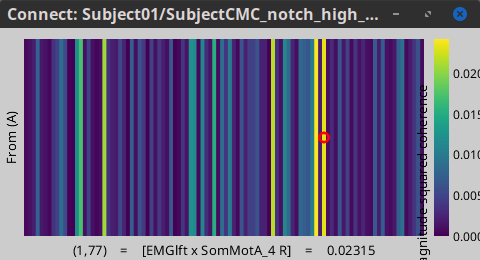
Open the '''Pipeline editor''': * Add the process '''Connectivity > Coherence AxB [2023]''' with the following parameters: * '''Time window''' = '''All file''' * '''Source channel (A)''' = `EMGlft` * Check '''Use scouts (B)''' * From the menu at the right, select '''Schaefer_100_17net''' * Select all the scouts * '''Scout function: Apply''': `before` * '''Scout function''': `mean` * Select '''Magnitude squared coherence''' as '''Connectivity metric''' * Select '''Fourier transform''' as '''Time-freq decomposition''' method * Click in the '''[Edit]''' button to set the parameters of the Fourier transform: * Select '''Matlab's FFT defaults''' as '''Frequency definition''' * '''FT window length''' = `0.5 s` * '''FT window overlap''' = `50%` * '''Highest frequency of interest''' = `80 Hz` * '''Time resolution''' = '''None''' * '''Estimate & save''' = '''across combined files/epochs''' * Add the process '''File > Add tag''' with the following parameters: * '''Tag to add''' = `(surface)(Constr)` * Select '''Add to file name''' * Run the pipeline || {{attachment:pro_coh_srfc_bef_sct.png}} || || {{attachment:pro_coh_srfc_bef_sct2.png}} || Double-click on the new file: the coherence spectra are not displayed on the cortex; they are plotted for each scout. The 1xN connectivity file can also be shown as an image as displayed below: || {{attachment:res_coh_srfc_bef_sct.png}} || || {{attachment:res_coh_srfc_bef_sct2.png}} || Note that at 14.65 Hz, the two highest peaks correspond to the '''SomMotA_4 R''' and '''SomMotA_2 R''' scouts, both located over the right primary motor cortex. {{{#!wiki caution The brain parcellation scheme is the user's decision. We recommend the Schaefer100 atlas used here by default. }}} == Coherence Scouts x Scouts == When computing whole-brain connectomes (i.e., NxN connectivity matrices between all brain sources), additional technical considerations need to be taken into account: the number of time series is typically too large to be computationally tractable on most workstations (see [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#Source-level_analyses|introduction]] above); if using unconstrained source maps, with 3 time series at each brain location, this requires an extra step to derive connectivity estimates. The last two sections explained the computation of coherence measures between one reference sensor (EMG) and brain source maps, with [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence#Method|sensor x sources]] and [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence#Coherence:_EMG_x_Scouts|sensor x scouts]] scenarios, both with constrained and unconstrained source orientation models. This section explains dimension reduction (across number of sources) using '''scouts''' to compute '''scouts x scouts''' coherence, for '''constrained''' and '''unconstrained''' source maps. As in the [[sensor-scout coherence]] scenario, it is necessary to specify two parameters to indicate how the data is aggregated in each scout: 1. The [[https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#Scout_function|scout function]] (for instance, the mean), and 2. When the scout aggregation procedure if performed, either '''before''' or '''after''' the coherence computation). The sections below explain the processing steps for the cases of '''constrained''' and '''unconstrained''' sources, for coherence between two scouts (''Scout-1'' and ''Scout-2''). In either case, the outcome is one coherence spectrum per pair of scouts. === Method for constrained sources === The computation steps depend of '''when''' the scout function is applied: * If the scout function is applied '''before''': The scout function (e.g., mean) is applied for all the source time series in each scout; resulting in '''1''' time series per scout. These scout time series are then used to compute coherence resulting in '''1''' coherence spectrum per scout. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_bef.png|{{attachment:diagram_1xn_coh_sct_bef.png||width="100%"}}]] * If the scout function applied '''after''': coherence is first computed between each combination of souces-scout1 and sources-scout2 resulting in '''N''' coherence spectra, with '''N''' being the product of sources-scout1 and sources-scout2. Finally the scout function is applied twice (once for each scout) to the resulting coherence spectra to obtain one coherence spectrum per scout pair. This is a considerably more greedy option that the '''"before"''' option above, as it computes coherence measures each source in scout1 and each source in scout2. This requires more computing resources; see [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#Source-level|introduction]] for an example. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_1xn_coh_sct_aft.png|{{attachment:diagram_1xn_coh_sct_aft.png||width="100%"}}]] === Method for unconstrained sources === As [[XX|described above]], when computing coherence with '''unconstrained sources''' it is necessary to perform dimension reduction across directions. When this reduction is done by first '''flattening''' the unconstrained sources, is performed on '''unconstrained sources''' coherence computations is carried out as described in the [[XX|previous section]]. However, if the sources are not flattened before computing coherence is carried out as follows: '''Before''': The scout function (e.g., mean) is applied on each direction on the elementary source time series each scout, which results in '''3''' time series per elementary direction, per scout. These scouts time series are then used to compute coherence measures, yielding to '''9''' coherence spectra (''1x-2x'', ''1x-2y'', ''1x-2z'', ''1y-2x'', ..., ''1z-2z'') for each pair of scouts. The resulting coherence spectra are finally aggregated across the '''9''' dimensions with maximum per frequency bin, resulting in one single coherence spectrum for each pair of scouts. [[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_nxn_coh_sct_bef.png|{{attachment:diagram_nxn_coh_sct_bef_small.gif||width="100%"}}]] '''After''': With this option, coherence is first computed between each pair of source locations (each with 3 time series) in the two scouts, yielding to '''9''' coherence spectra for each pair of sources in the scouts. Then, these coherence spectra are aggregated across the 9 combinations of dimensions (''1x-2x'', ''1x-2y'', ''1x-2z'', ''1y-2x'', ..., ''1z-2z'') to obtain one single coherence spectrum for each pair of sources within the scouts. Finally, the '''scout function''' (e.g., mean) is applied twice (once per scout) to the coherence spectra for each source within the scouts to obtain one single coherence spectrum for each pair of scouts. Applying the scout function '''"after"''' is a considerably more greedy option that the applying the scout function '''"before"''', as it computes coherence measures between all 45000x45000 elementary source signals, and this number is multiplied by the number of frequency bins. This requires more computing resources; see the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Connectivity#Source-level_analyses|introduction]] for an example.[[https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_nxn_coh_sct_aft.png|{{attachment:diagram_nxn_coh_sct_aft_small.gif|https://neuroimage.usc.edu/brainstorm/Tutorials/CorticomuscularCoherence?action=AttachFile&do=get&target=diagram_nxn_coh_sct_aft.png|width="100%"}}]] === Example === Here, we will compute the coherence from scouts, using '''mean''' as scout function with the '''before''' option. We will use the [[https://www.biorxiv.org/content/biorxiv/early/2017/07/16/135632.full.pdf|Schaefer 100 parcellation]] atlas on to the orientation-constrained source map. * Use Search Database (<<Icon(iconZoom.gif)>>) to select the '''Left''' trials with their respective '''(surface)(Constr)''' source maps, as shown in the [[#Coherence_1xN_.28source_level.29|previous section]]. * In the Process2: '''Left '''trial group into both the '''Files A''' and '''Files B''' boxes. Select '''Process recordings''' (<<Icon(iconEegList.gif)>>) for Files A, and '''Process sources''' (<<Icon(iconResultList.gif)>>) for Files B. . {{attachment:process2.png||width="80%"}} Open the '''Pipeline editor''': * Add the process '''Connectivity > Coherence AxB [2023]''' with the following parameters: * '''Time window''' = '''All file''' * '''Source channel (A)''' = `EMGlft` * Check '''Use scouts (B)''' * From the menu at the right, select '''Schaefer_100_17net''' * Select all the scouts * '''Scout function: Apply''': `before` * '''Scout function''': `mean` * Select '''Magnitude squared coherence''' as '''Connectivity metric''' * Select '''Fourier transform''' as '''Time-freq decomposition''' method * Click in the '''[Edit]''' button to set the parameters of the Fourier transform: * Select '''Matlab's FFT defaults''' as '''Frequency definition''' * '''FT window length''' = `0.5 s` * '''FT window overlap''' = `50%` * '''Highest frequency of interest''' = `80 Hz` * '''Time resolution''' = '''None''' * '''Estimate & save''' = '''across combined files/epochs''' * Add the process '''File > Add tag''' with the following parameters: * '''Tag to add''' = `(surface)(Constr)` * Select '''Add to file name''' * Run the pipeline || {{attachment:pro_coh_srfc_bef_sct.png}} || || {{attachment:pro_coh_srfc_bef_sct2.png}} || Double-click on the new file: the coherence spectra are not displayed on the cortex; they are plotted for each scout. The 1xN connectivity file can also be shown as an image as displayed below: || {{attachment:res_coh_srfc_bef_sct.png}} || || {{attachment:res_coh_srfc_bef_sct2.png}} || Note that at 14.65 Hz, the two highest peaks correspond to the '''SomMotA_4 R''' and '''SomMotA_2 R''' scouts, both located over the right primary motor cortex. {{{#!wiki caution The brain parcellation scheme is the user's decision. We recommend the Schaefer100 atlas used here by default. }}} === Method for mixed source models === [[https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas|Mixed models]] refer to source models with multiple regions that may include '''surface''' and '''volume''' regions, and regions with different source orientation constraints. To compute connectivity between surface and volume regions, you must first [[https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas#Volume_scouts|create volume scouts]] for the volume regions of the mixed model, and then use the process2 tab with the same files on both sides. On one side you can select surface scouts, and the other, the volume scouts you created. For connectivity between surface regions only, or volume regions only, use the process1 tab. <<TAG(Advanced)>> |
| Line 335: | Line 634: |
| * Kilner JM, Baker SN, Salenius S, Hari R, Lemon RN. <<BR>> [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Human Cortical Muscle Coherence Is Directly Related to Specific Motor Parameters]]. <<BR>> J Neurosci. 2000 Dec 1;20(23):8838–45. '' '' * ''Liu J, Sheng Y, Liu H. <<BR>> [[https://doi.org/10.3389/fnhum.2019.00100Corticomuscular%20Coherence%20and%20Its%20Applications:%20A%20Review|https://doi.org/10.3389/fnhum.2019.00100Corticomuscular%20Coherence%20and%20Its%20Applications:%20A%20Review]]. Front Hum Neurosci. 2019 Mar 20;13:100. '' |
* Kilner JM, Baker SN, Salenius S, Hari R, Lemon RN. <<BR>> [[https://doi.org/10.1523/JNEUROSCI.20-23-08838.2000|Human Cortical Muscle Coherence Is Directly Related to Specific Motor Parameters]]. <<BR>> J Neurosci. 2000 Dec 1;20(23):8838–45. * Liu J, Sheng Y, Liu H. <<BR>> [[https://doi.org/10.3389/fnhum.2019.00100|Corticomuscular Coherence and Its Applications: A Review]]. <<BR>> Front Hum Neurosci. 2019 Mar 20;13:100. * Sadaghiani S, Brookes MJ, Baillet S. <<BR>> [[https://doi.org/10.1016/j.neuroimage.2021.118788|Connectomics of human electrophysiology]]. <<BR>> NeuroImage. 2022 Feb;247:118788. |
| Line 340: | Line 641: |
| * ''Tutorial: [[Tutorials/TutVolSource|Volume source estimation]] '' ==== Forum discussions ==== * ''Forum: Minimum norm units (pA.m): [[http://neuroimage.usc.edu/forums/showthread.php?1246-Doubt-about-current-density-units-pA.m-or-pA-m2|http://neuroimage.usc.edu/forums/showthread.php?1246]] '' ''<<HTML(<!-- END-PAGE -->)>> '' ''<<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/CorticomuscularCoherence)>> '' |
* Tutorial: [[Tutorials/Connectivity|Functional connectivity]] * Tutorial: [[Tutorials/SourceEstimation|Source estimation]] * Tutorial: [[Tutorials/TutVolSource|Volume source estimation]] * Tutorial: [[Tutorials/Scouts|Scouts]] * Tutorial: [[Tutorials/ConnectivityGraph|Connectivity graphs]] == Scripting == The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_coherence.m|brainstorm3/toolbox/script/tutorial_coherence.m]] <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:500px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("https://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_coherence.m")>><<HTML(</div >)>> <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/CorticomuscularCoherence)>> |
Corticomuscular coherence (CTF MEG)
Authors: Raymundo Cassani, Francois Tadel, Sylvain Baillet.
Corticomuscular coherence measures the degree of similarity between electrophysiological signals (MEG, EEG, ECoG sensor traces or source time series, especially over the contralateral motor cortex) and the EMG signals recorded from muscle activity during voluntary movements. Cortical-muscular signals similarities are conceived as due mainly to the descending communication along corticospinal pathways between primary motor cortex (M1) and the muscles attached to the moving limb(s). For consistency purposes, the present tutorial replicates, with Brainstorm tools, the processing pipeline "Analysis of corticomuscular coherence" of the FieldTrip toolbox.
Contents
Dataset description
The dataset is identical to that of the FieldTrip tutorial: Analysis of corticomuscular coherence:
- One participant,
- MEG recordings: 151-channel CTF MEG system,
- Bipolar EMG recordings: from left and right extensor carpi radialis longus muscles,
- EOG recordings: used for detection and attenuation of ocular artifacts,
- MRI: 1.5T Siemens system,
- Task: The participant lifted their hand and exerted a constant force against a lever for about 10 seconds. The force was monitored by strain gauges on the lever. The participant performed two blocks of 25 trials using either their left or right hand.
- Here we describe relevant Brainstorm tools via the analysis of the left-wrist trials. We encourage the reader to practice further by replicating the same pipeline using right-wrist trials!
Corticomuscular coherence:
Coherence measures the linear relationship between two signals in the frequency domain.
Previous studies (Conway et al., 1995, Kilner et al., 2000) have reported corticomuscular coherence effects in the 15–30 Hz range during maintained voluntary contractions.
- TODO: IMAGE OF EXPERIMENT, SIGNALS and COHERENCE
Download and installation
Pre-requisites
Please make sure you have completed the get-started tutorials, prior to going through the present tutorial.
- Have a working copy of Brainstorm installed on your computer.
Install the CAT12 Brainstorm plugin, to perform MRI segmentation from the Brainstorm dashboard.
Download the dataset
Download SubjectCMC.zip from the FieldTrip download page:
https://download.fieldtriptoolbox.org/tutorial/SubjectCMC.zip- Unzip the downloaded archive file in a folder outside the Brainstorm database or app folders (for instance, directly on your desktop).
Brainstorm
- Launch Brainstorm (via the Matlab command line or the Matlab-free stand-alone version of Brainstorm).
Select from the menu File > Create new protocol. Name the new protocol TutorialCMC and select the options:
No, use individual anatomy,
No, use one channel file per acquisition run.
Importing anatomy
Right-click on the newly created TutorialCMC node > New subject > Subject01.
Keep the default options defined for the study (aka "protocol" in Brainstorm's vernacular).Switch to the Anatomy view of the protocol (
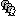 ).
). Right-click on the Subject01 > Import MRI:
Select the file format: All MRI files (subject space)
Select the file: SubjectCMC/SubjectCMC.mri
This step will launch Brainstorm's MRI viewer, where coronal, sagittal and axial cross-sections of the MRI volume are displayed. note that three anatomical fiducials (left and right pre-auricular points (LPA and RPA), and nasion (NAS)) are automatically identified. These fiducials are located near the left/right ears and just above the nose, respectively. Click on Save.
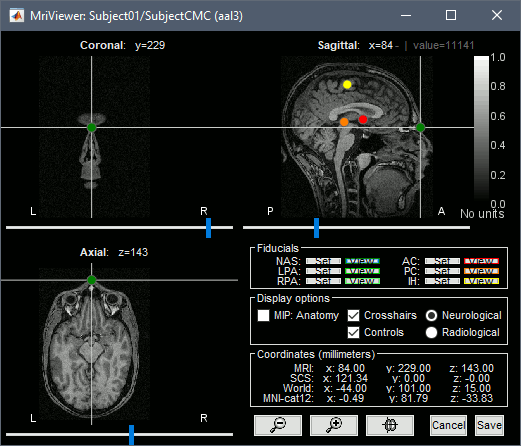
In all typical Brainstorm workflows from this tutorial handbook, we recommend processing the MRI volume at this stage, before importing the functional (MEG/EEG) data. However, we will proceed differently, for consistency with the original FieldTrip pipeline, and readily obtain sensor-level coherence results. We will proceed with MRI segmentation below, before performing source-level analyses.
- We still need to verify the proper geometric registration (alignment) of MRI with MEG. We will therefore now extract the scalp surface from the MRI volume.
Right-click on the MRI (
 ) > MRI segmentation > Generate head surface.
) > MRI segmentation > Generate head surface.
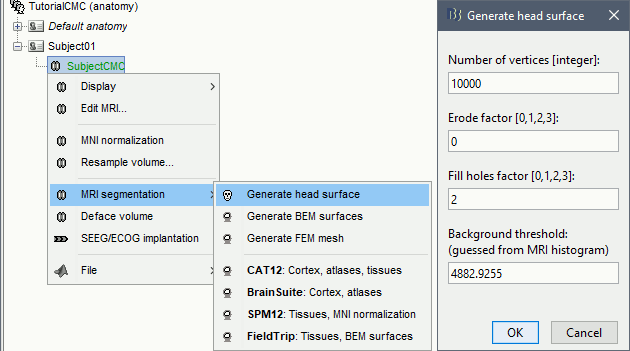
Double-click on the newly created surface to display the scalp in 3-D.
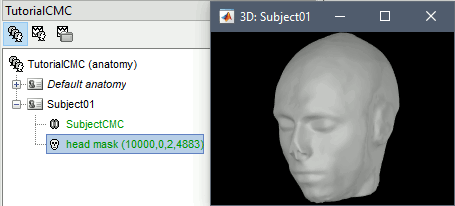
MEG and EMG recordings
Link the recordings
Switch now to the Functional data view of your database contents (
 ).
). Right-click on Subject01 > Review raw file:
Select the appropriate MEG file format: MEG/EEG: CTF(*.ds; *.meg4; *.res4)
Select the data file: SubjectCMC.ds
A new folder SubjectCMC is created in the Brainstorm database explorer. Note the "RAW" tag over the icon of the folder (
 ), indicating that the MEG files contain unprocessed, continuous data. This folder includes:
), indicating that the MEG files contain unprocessed, continuous data. This folder includes: CTF channels (191) is a file with all channel information, including channel types (MEG, EMG, etc.), names, 3-D locations, etc. The total number of channels available (MEG, EMG, EOG etc.) is indicated between parentheses.
Link to raw file is a link to the original data file. Brainstorm reads all the relevant metadata from the original dataset and saves them into this symbolic node of the data tree (e.g., sampling rate, number of time samples, event markers). As seen elsewhere in the tutorial handbook, Brainstorm does not create copies by default of (potentially large) unprocessed data files (more information).
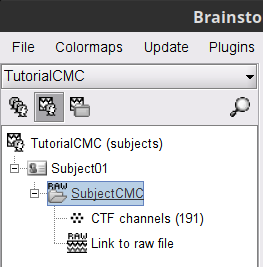
MEG-MRI coregistration
This registration step is to align the MEG coordinate system with the participant's anatomy from MRI (more info). Here we will use only the three anatomical landmarks stored in the MRI volume and specific at the moment of MEG data collection. From the FieldTrip tutorial:
"To measure the head position with respect to the sensors, three coils were placed at anatomical landmarks of the head (nasion, left and right ear canal). [...] During the MRI scan, ear molds containing small containers filled with vitamin E marked the same landmarks. This allows us, together with the anatomical landmarks, to align source estimates of the MEG with the MRI."To visually appreciate the correctness of the registration, right-click on the CTF channels node > MRI registration > Check. This opens a 3-D figure showing the inner surface of the MEG helmet (in yellow), the head surface, the fiducial points and the axes of the subject coordinate system (SCS).
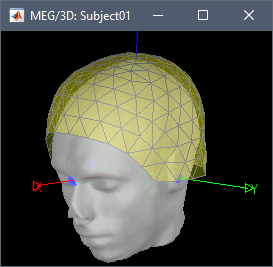
Reviewing
Right-click on the Link to raw file > Switch epoched/continuous to convert how the data i stored in the file to a continuous reviewing format, a technical detail proper to CTF file formatting.
Right-click on the Link to raw file > MEG > Display time series (or double-click on its icon). This will display data time series and enable the Time panel and the Record tab in the main Brainstorm window (see specific features in "explore data time series").
Right-click on the Link to raw file > EMG > Display time series.
Event markers
The colored dots at the top of the figure, above the data time series, indicate event markers (or triggers) saved along with MEG data. The actual onsets of the left- and right-wrist trials are not displayed yet: they are saved in an auxiliary channel of the raw data named Stim. To add these markers to the display, follow this procedure:
With the time series figure still open, go to the Record tab and select File > Read events from channel. Event channels = Stim, select Value, Reject events shorter than 12 samples. Click Run.
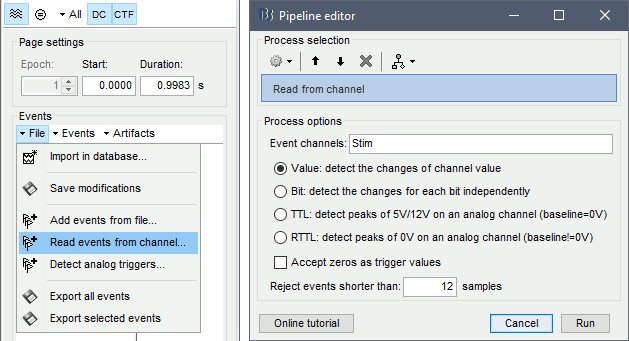
The rejection of short events is nececessary in this dataset, because transitions between values in the Stim channel may span over several time samples. Otherwise, and for example, an event U1 would be created at 121.76s because the transition from the event U1025 back to zero features two unwanted values at the end of the event. The rejection criteria is set to 12 time samples (10ms), because the duration of all the relevant triggers is longer than 15ms. This value is proper to each dataset, so make sure you verify trigger detections from your own dataset.
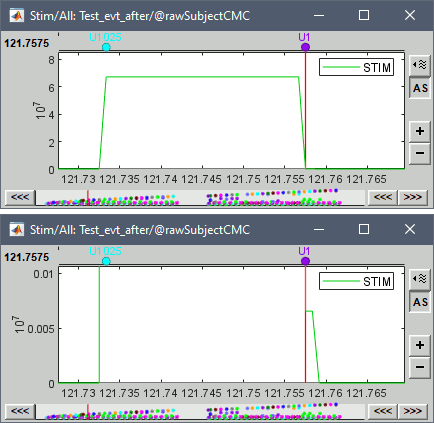
New event markers are created and now shown in the Events section of the tab, along with previous event categories. In this tutorial, we will only use events U1 through U25, which correspond to the beginning of each of the 25 left-wrist trials of 10 seconds. Following FieldTrip's tutorial, let's reject trial #7, event U7.
Delete all unused events: Select all the events except U1-U6 and U8-25 (Ctrl+click / Shift+click), then menu Events > Delete group (or press the Delete key).
Merge events: Select all the event groups, then select from the menu Events > Merge group > "Left". A new event category called "Left" now indicate the onsets of 24 trials of left-wrist movements.
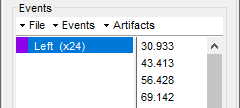
Pre-processing
In this tutorial, we will analyze only the Left trials (left-wrist extensions). In the following sections, we will process only the first 330 s of the original recordings, where the left-wrist trials were performed.
Power line artifacts
In the Process1 box: Drag and drop the Link to raw file.
Run process Frequency > Power spectrum density (Welch):
Time window: 0-330 s
Window length: 10 s
Overlap: 50%
Sensor types: MEG, EMG
Double-click on the new PSD file to visualize the power spectrum density of the data.
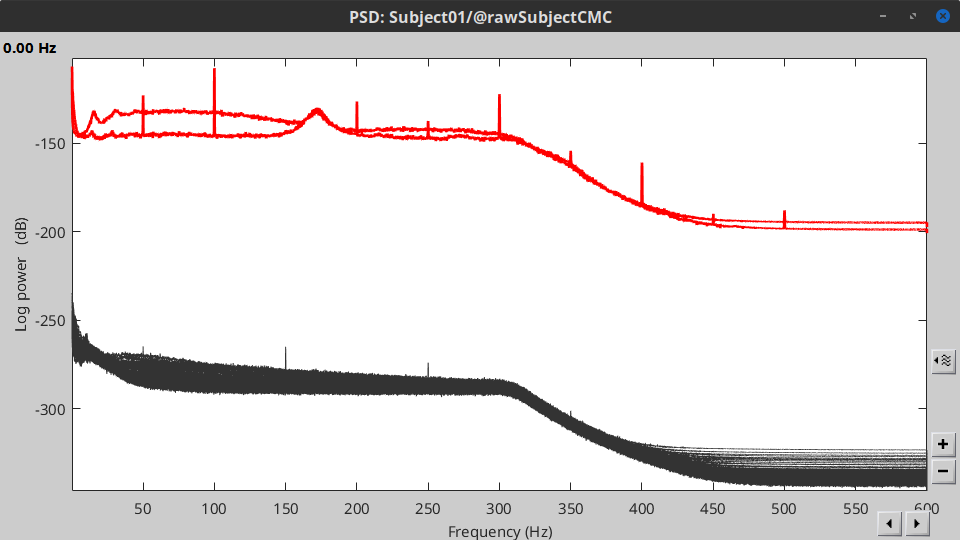
- The PSD plot shows two groups of sensors: EMG (highlighted in red above) and the MEG spectra below (black lines). Peaks at 50Hz and harmonics (100, 150, 200Hz and above) indicate the European power line frequency and are clearly visible. We will now use notch filters to attenuate power line contaminants at 50, 100 and 150 Hz.
In the Process1 box: Drag and drop the Raw | clean node.
Run the process Pre-processing > Notch filter with:
Check Process the entire file at once
Sensor types: MEG, EMG
Frequencies to remove (Hz): 50, 100, 150
In case you get a memory error message:
These MEG recordings have been saved before applying the CTF 3rd-order gradient compensation, for noise reduction. The compensation weights are therefore applied on the fly when Brainstorm reads data from the file. However, this requires reading all the channels at once. By default, the frequency filter are optimized to process channel data sequentially, which is incompatible with applying the CTF compensation on the fly. This setting can be overridden with the option Process the entire file at once, which requires loading the entire file in memory at once, which may crash teh process depending on your computing resources (typically if your computer's RAM < 8GB). If this happens: run the process Artifacts > Apply SSP & CTF compensation on the file first, then rune the notch filter process without the option "Process the entire file at once" (more information).A new folder named SubjectCMC_clean_notch is created. Obtain the PSD of these data to appreciate the effect of the notch filters. As above, please remember to indicate a Time window restricted from 0 to 330 s in the options of the PSD process.
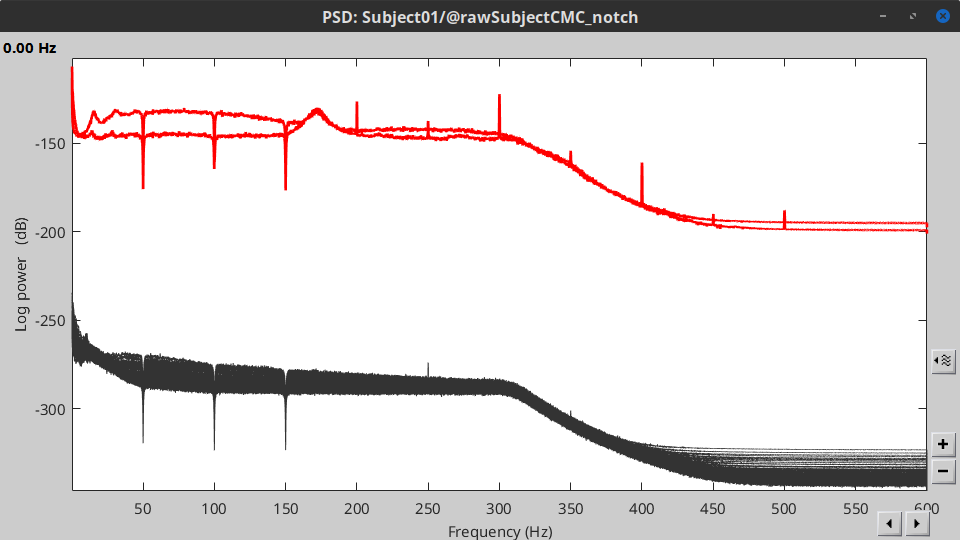
EMG: Filter and rectify
Two typical pre-processing steps for EMG consist in high-pass filtering and rectifying.
In the Process1 box: drag and drop the Raw | notch(50Hz 100Hz 150Hz) node.
Add the process Pre-process > Band-pass filter
Sensor types = EMG
Lower cutoff frequency = 10 Hz
Upper cutoff frequency = 0 Hz
Add the process Pre-process > Absolute values
Sensor types = EMG
- Run the pipeline
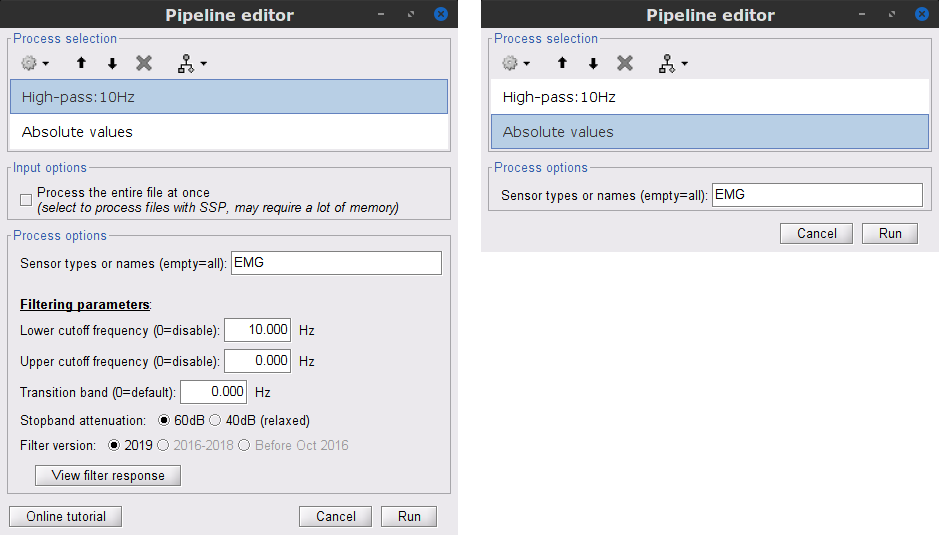
Delete intermediate files that won't be needed anymore: Select folders SubjectCMC_notch and SubjectCMC_notch_high, then press the Delete key (or right-click > File > Delete).
MEG: Blink SSP and bad segments
Stereotypical artifacts such eye blinks and heartbeats can be identified from their respective characteristic spatial distributions. Their contamination of MEG signals can then be attenuated specifically using Signal-Space Projections (SSPs). For more details, consult the specific tutorial sections about the detection and removal of artifacts with SSP. The present tutorial dataset features an EOG channel but no ECG. We will therefore only remove artifacts caused by eye blinks.
Blink correction with SSP
Right-click on the pre-processed file > MEG > Display time series and EOG > Display time series.
In the Record tab: Artifacts > Detect eye blinks, and use the parameters:
Channel name= EOG
Time window = 0 - 330 s
Event name = blink
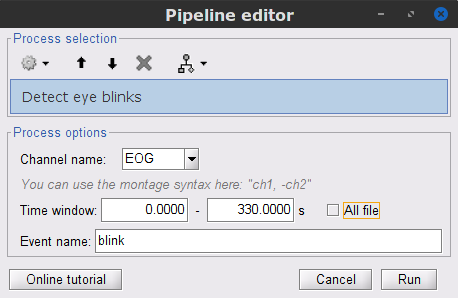
Three categories of blink events are created. Review the traces of the EOG channels around a few of these events to ascertain they are related to eye blinks. In the present case, we note that the blink group contains genuine eye blinks, and that groups blink2 and blink3 capture saccades.
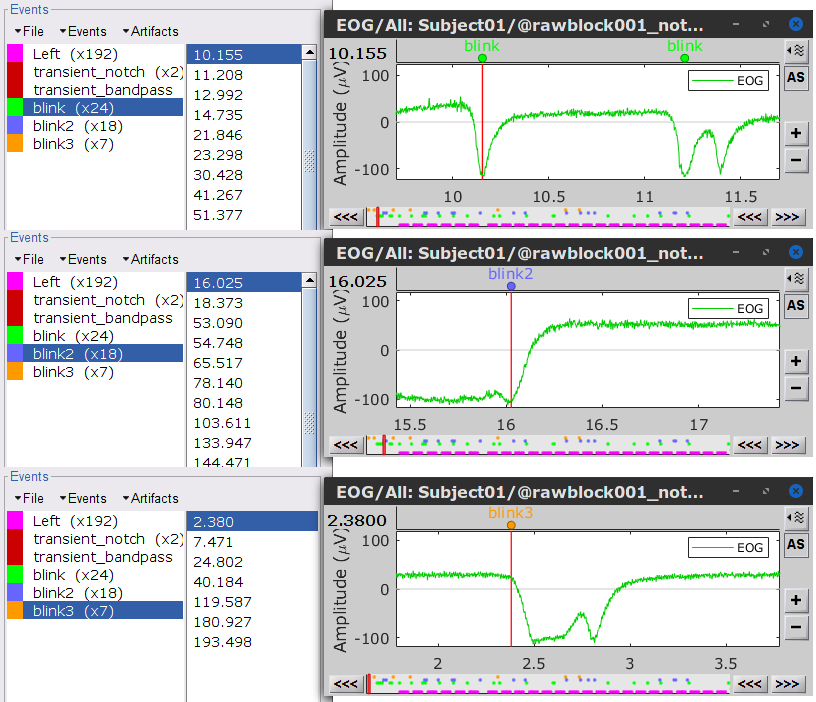
To remove blink artifacts with SSP, go to Artifacts > SSP: Eye blinks:
Event name=blink
Sensors=MEG
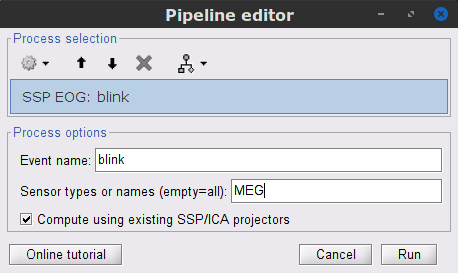
Display the time series and topographies of the first two SSP components identified. In the present case, only the first SSP component can be clearly related to blinks: the percentage of variance explained is substantially higher than the other compoments', the spatial topography of the component is also typical of eye blinks, and the corresponding time series is similar to the EOG signal around blinks. Select only component #1 for removal.
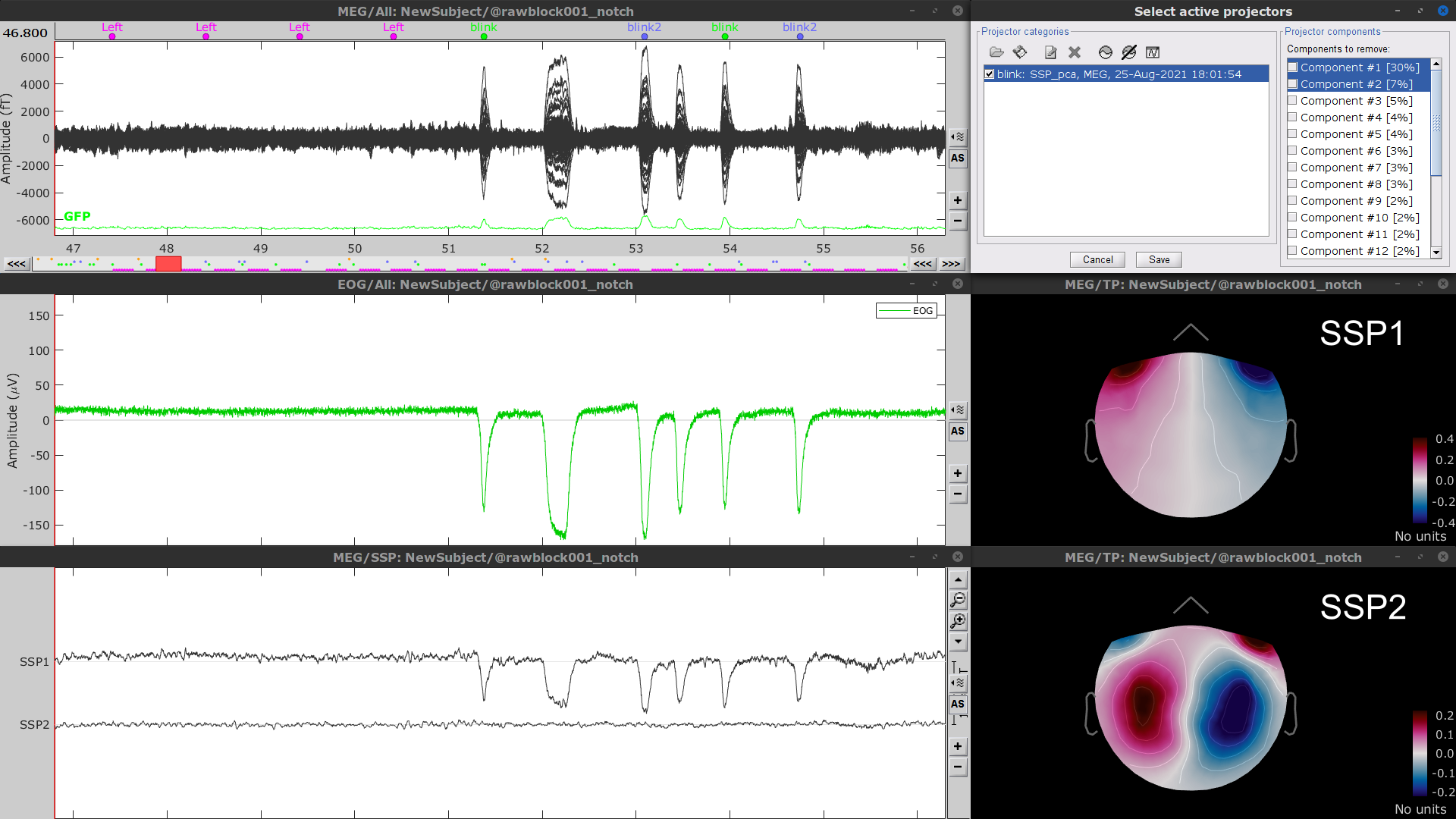
Close all visualization figures by clicking on the large × at the top-right of the main Brainstorm window.
Detection of "bad" data segments
Here we will use the automatic detection of artifacts to identify data segments contaminated by e.g., large eye and head movements, or muscle contractions.
Display the MEG and EOG time series. In the Record tab, select Artifacts > Detect other artifacts and enter the following parameters:
Time window = 0 - 330 s
Sensor types=MEG
Sensitivity=3
Check both frequency bands 1-7 Hz and 40-240 Hz
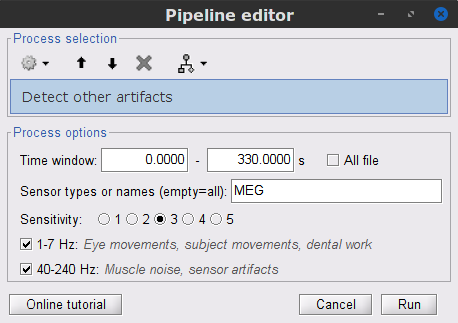
- You are encouraged to review all the segments marked using this procedure. With the present data, all marked segments do contain clear artifacts.
Select the 1-7Hz and 40-240Hz event groups and select Events > Mark group as bad. Alternatively, you can add the prefix bad_ to the event names. Brainstorm will automatically discard these data segments from further processing.
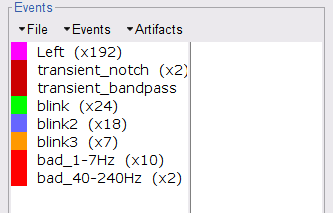
- Close all visualization windows and reply "Yes" to the save the modifications query.
Epoching
We are now finished with the pre-processing of EMG and MEG recordings. We will now extract and import specific data segments of interest into the Brainstorm database for further derivations. As mentioned previously, we will focus on the Left category of events (left-wrist movements). For consistency with the FieldTrip tutorial, we will analyze 8s of recordings following each movement (from the original 10s around each trial), and split them in 1-s epochs. For each 1-s epoch, the DC offset will be also removed from each MEG channel. And finally, we will uniform the time vector for all 1-s epochs. This last steps is needed as the time vector of each 1-s vector is with respect to the event that was used to import the 8-s trial.
- In the Process1 box: Drag-and-drop the pre-processed file.
Select the process Import > Import recordings > Import MEG/EEG: Events:
Subject name = Subject01
Folder name = empty
Event names = Left
Time window = 0 - 330 s
Epoch time = 0 - 8000 ms
Split recordings in time blocks = 1 s
Uncheck Create a separate folder for each event type
Check Ignore shorter epochs
Check Use CTF compensation
Check Use SSP/ICA projectors
Add the process Pre-process > Remove DC offset:
Baseline = All file
Sensor types = MEG
Check Overwrite input files
Add the process Standardize > Uniform epoch time:
Interpolation method = spline
Check Overwrite input files
- Run the pipeline
|
|
|
A new folder SubjectCMC_notch_high_abs without the 'raw' indication is now created, which includes 192 individual epochs (24 trials x 8 1-s epochs each). The epochs that overlap with a "bad" event are also marked as bad, as shown with an exclamation mark in a red circle (
 ). These bad epochs will be automatically ignored by the Process1 and Process2 tabs, and from all further processing.
). These bad epochs will be automatically ignored by the Process1 and Process2 tabs, and from all further processing. 
Comparison with FieldTrip
The figures below show the EMG and MRC21 channels (a MEG sensor over the left motor-cortex) from the epoch #1.1, in Brainstorm (left), and from the FieldTrip tutorial (right).
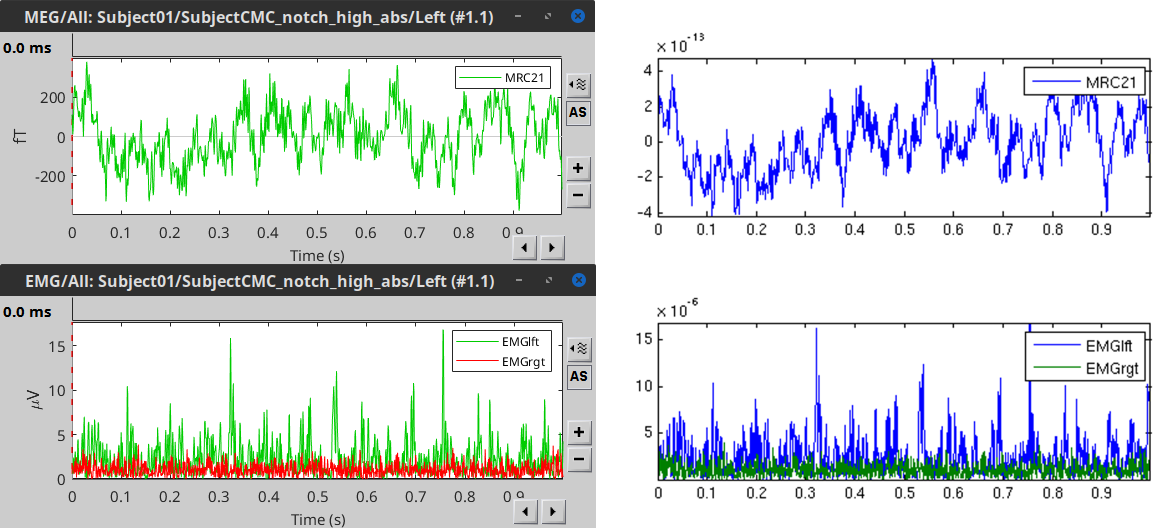
Coherence: EMG x MEG
Let's compute the magnitude-squared coherence (MSC) between the left EMG and the MEG channels.
In the Process1 box, drag and drop the Left (192 files) trial group.
Select the process Connectivity > Coherence 1xN [2023]:
Time window = Check All file
Source channel = EMGlft
Sensor types = MEG
Do not check Include bad channels
Select Magnitude squared coherence as Connectivity metric
Select Fourier transform as Time-freq decomposition method
Click in the [Edit] button to set the parameters of the Fourier transform:
Select Matlab's FFT defaults as Frequency definition
FT window length = 0.5 s
FT window overlap = 50%
Highest frequency of interest = 80 Hz
Time resolution = None
Estimate & save = across combined files/epochs
More details on the Coherence process can be found in the Connectivity Tutorial.
Add the process File > Add tag with the following parameters:
Tag to add = MEG sensors
Select Add to file name
- Run the pipeline:
|
|
|
Double-click on the resulting data node MSCoh-stft: EMGlft (185 files) | MEG sensors to display the MSC spectra. Click on the maximum peak in the 15 to 20 Hz range, and press Enter to display the spectrum from the selected sensor in a new window. This spectrum is that of channel MRC21, and shows a prominent peak at 17.58 Hz. Use the frequency slider (under the Time panel) to explore the MSC output across frequencies.
Right-click on the spectrum and select 2D Sensor cap for a topographical representation of the magnitude of the coherence results across the sensor array. You may also use the shortcut Ctrl-T. The sensor locations can be displayed with a right-click and by selecting Channels > Display sensors from the contextual menu (shortcut Ctrl-E).
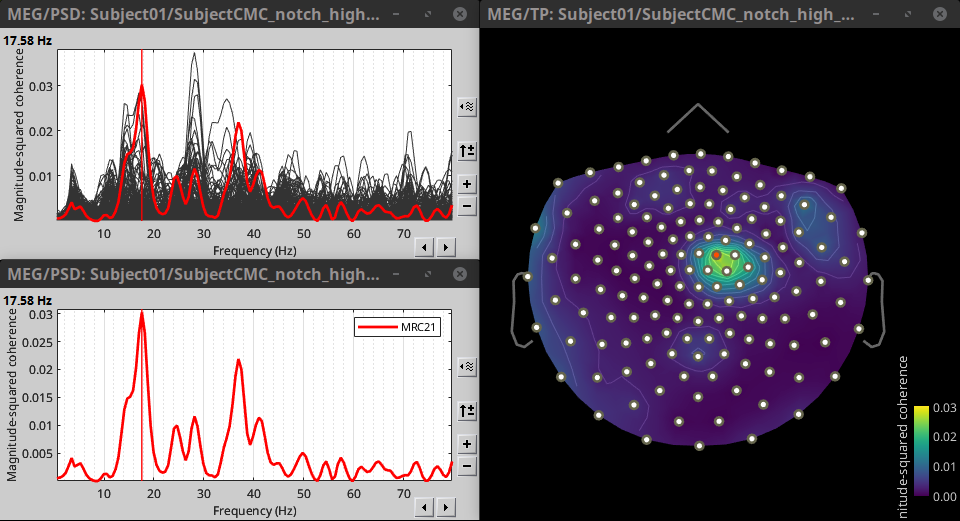
We can now average the magnitude of the MSC across the beta band (15-20 Hz).
In the Process1 box, select the new mscohere file.Run process Frequency > Group in time or frequency bands:
Select Group by frequency bands
Type cmc_band / 15, 20 / mean in the text box.
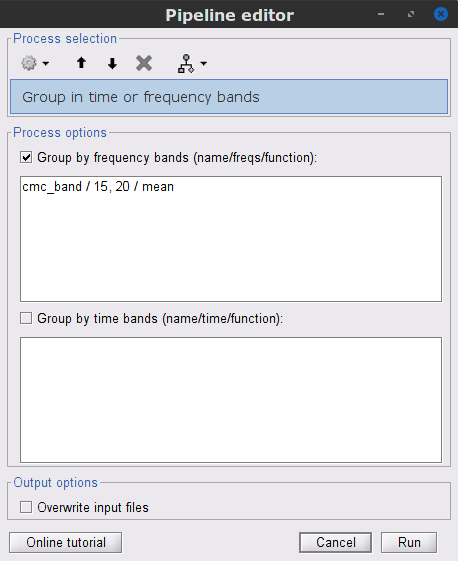
The resulting MSCoh-stft:...|tfbands node contains one MSC value for each sensor (the MSC average in the 15-20 Hz band). Right-click on the file to display the 2D or 3D topography of the MSC beta-band measure.
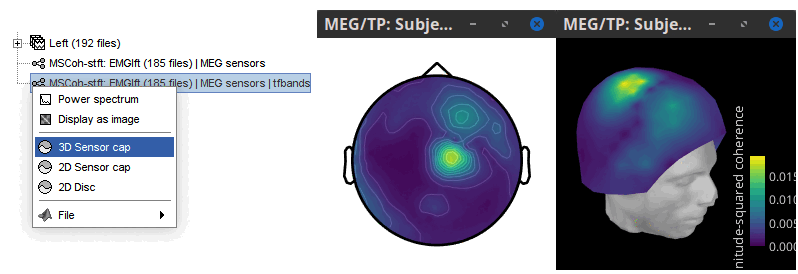
Higher MSC values the EMG signal and MEG sensor signals map over the contralateral set of central sensors in the beta band. Sensor-level connectivity can be ambiguous to interpret anaotmically though. We will now map the magnitude of EMG-coherence across the brain (MEG sources).
Source estimation
MRI segmentation
We first need to extract the cortical surface from the T1 MRI volume we imported at the beginning of this tutorial. CAT12 is a Brainstorm pluing that will perform this task in 30-60min.
Switch back to the Anatomy view of the protocol (
 ).
). Right-click on the MRI (
 ) > MRI segmentation > CAT12:
) > MRI segmentation > CAT12: Number of vertices: 15000
Anatomical parcellations: Yes
Cortical maps: No
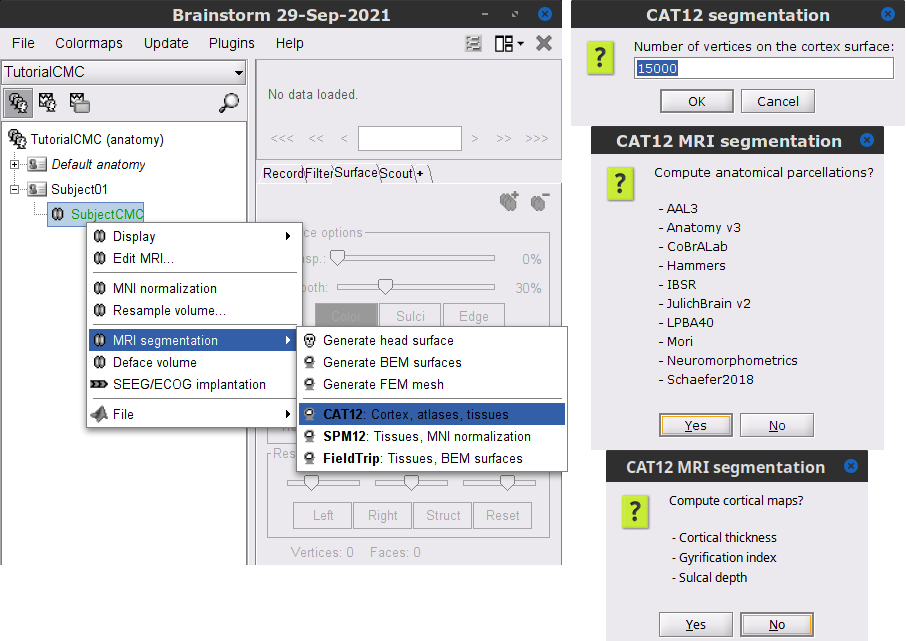
Keep the low-resolution central surface selected as the default cortex (central_15002V). This surface is the primary output of CAT12, and is shown half-way between the pial envelope and the grey-white interface (more information). The head surface was recomputed during the process and duplicates the previous surface obtained above: you can either delete one of the head surfaces or ignore this point for now.
For quality control, double-click on the head and central_15002V surfaces to visualize them in 3D.
Head models
We will perform source modeling using a distributed model approach for two different source spaces: the cortex surface and the entire MRI volume. Forward models are called head models in Brainstorm. They account for how neural electrical currents produce magnetic fields captured by sensors outside the head, considering head tissues electromagnetic properties and geometry, independently of actual empirical measurements (more information). A distinct head model is required for the cortex surface and head volume source spaces.
Cortical surface
Go back to the Functional data view of the database.
Right-click on the channel file of the imported epoch folder > Compute head model.
Comment = Overlapping spheres (surface)
Source space = Cortex surface
Forward model = MEG Overlapping spheres.
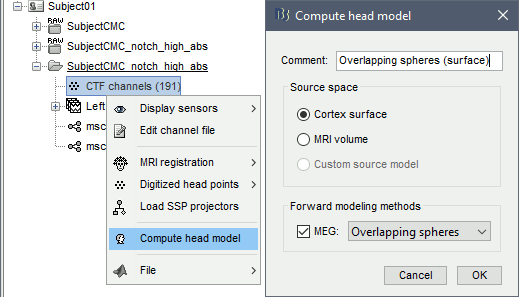
Whole-head volume
Right-click on the channel file again > Compute head model.
Comment = Overlapping spheres (volume)
Source space = MRI volume
Forward model = Overlapping spheres.
Select Regular grid and Brain
Grid resolution = 5 mm
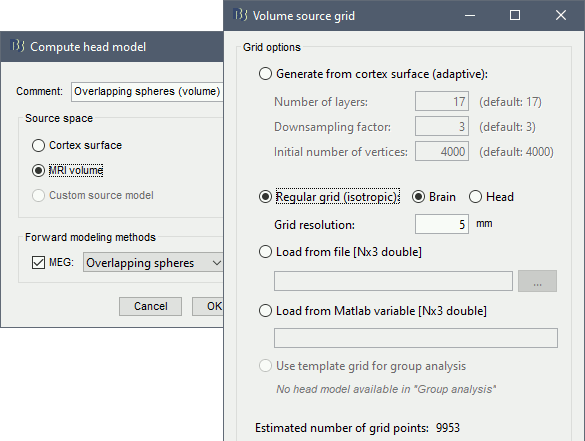
The Overlapping spheres (volume) head model is now added to the database explorer. The green color of the name indicates this is the default head model for the current folder: you can decide to use another head model available by double clicking on its name.
Noise covariance
The recommendation for MEG source imaging is to extract basic noise statistics from empty-room recordings. When not available, as here, resting-state data can be used as proxies for MEG noise covariance. We will use a segment of the MEG recordings, away from the task and major artifacts: 18s-29s.
Right-click on the clean continuous file > Noise covariance > Compute from recordings.
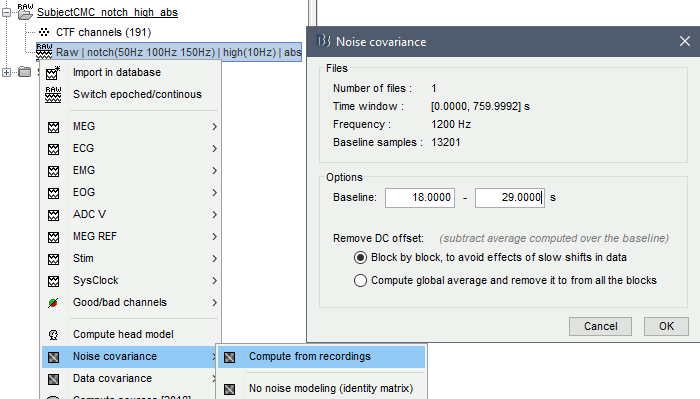
Right-click on the Noise covariance (
 ) > Copy to other folders.
) > Copy to other folders.
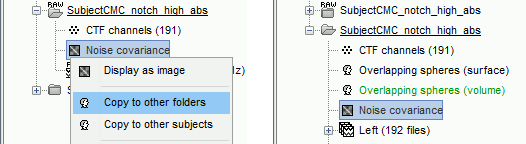
Inverse models
We will now compute three inverse models, with different source spaces: cortex surface with constrained dipole orientations (normal to the cortex), cortex surface with unconstrained orientation, and MRI volume (more information).
Cortical surface
Right-click on Overlapping spheres (surface) > Compute sources:
Minimum norm imaging
Current density map
Constrained: Normal to the cortex
Comment = MN: MEG (surface)
Repeat the previous step, but this time select Unconstrained in the Dipole orientations field.
|
|
|
Whole-head volume
Right-click on the Overlapping spheres (volume) > Compute sources:
Current density map
Unconstrained
Comment = MN: MEG (volume)
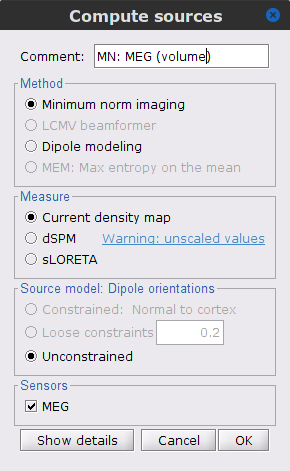
Three imaging kernels (
 ) are now available in the database explorer. Note that each trial is associated with three source links (
) are now available in the database explorer. Note that each trial is associated with three source links ( ). The concept of imaging kernels is explained in Brainstorm's main tutorial on source mapping.
). The concept of imaging kernels is explained in Brainstorm's main tutorial on source mapping. 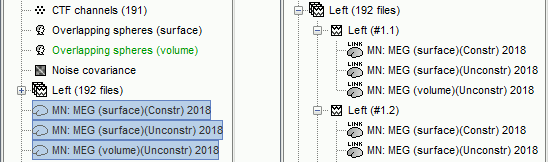
Coherence: EMG x Sources
We can now compute the coherence between the EMG signal and all brain source time series, for each of the source models considered. Let's start with the cortical surface/constrained model.
To select the source maps to include in the coherence estimation, click on the Search Database button (
 ), and select New search. Set the parameters as shown below, and click on Search.
), and select New search. Set the parameters as shown below, and click on Search. 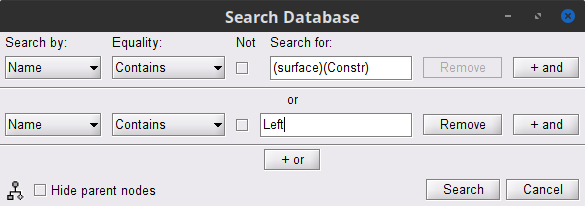
- This creates a new tab in the database explorer, showing only the files that match the search criteria.
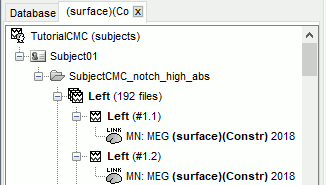
Click the Process2 tab at the bottom of the main Brainstorm window.
Files A: Drag-and-drop the Left (192 files) group, select Process recordings (
 ).
). Files B: Drag-and-drop the Left (192 files) group, select Process sources (
 ).
). - Objective: Extract from the same files both the EMG recordings (Files A) and the sources time series (Files B), then compute the coherence measure between these two categories of time series. Note that the blue labels over the file lists indicate that there are 185 "good" files (7 bad epochs).
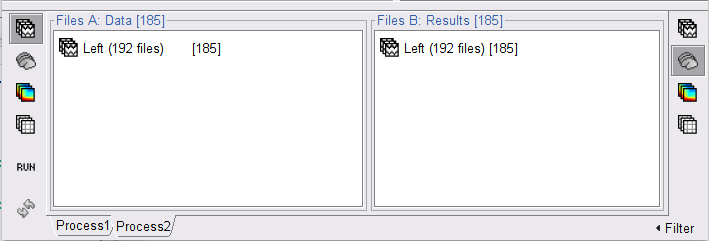
Select the process Connectivity > Coherence AxB [2023]:
Time window = All file
Source channel (A) = EMGlft
Uncheck Use scouts (B)
Check Flatten unconstrained source orientations
Click in the [Edit] button of the PCA options:
Select across all epochs/files as PCA method
PCA time window = All file
Check Remove DC offset
Baseline = All file
Ignore the Scout function options as we are not using scouts
Select Magnitude squared coherence as Connectivity metric
Select Fourier transform as Time-freq decomposition method
Click in the [Edit] button to set the parameters of the Fourier transform:
Select Matlab's FFT defaults as Frequency definition
FT window length = 0.5 s
FT window overlap = 50%
Highest frequency of interest = 80 Hz
Time resolution = None
Estimate & save = across combined files/epochs
Add the process File > Add tag:
Tag to add = (surface)(Constr)
Select Add to file name
- Run the pipeline
|
|
|
Repeat the steps above to compute the EMG-sources coherence for the other source models: surface/unconstrained and volume:
Edit the search criteria accordingly: Right-click on the search tab > Edit search.
- This updates automatically the file selection in the Process2 tab.
Select the processes as indicated above and update the file tag according to the type of sources.
The parameters Flatten unconstrained source orientations and PCA options did not have any impact on constrained sources, as there already consist on one time series per source location. However, when used with unconstrained sources, the three time series per source location are flattened into one before computing coherence.
If the option Flatten unconstrained source orientations is unchecked for unconstrained sources, coherence is computed between the sensor and each of the three time series per source location, resulting in three coherence spectra which are collapsed into one using the maximum value for each frequency band.
Close the search tab. If the 3 new connectivity files
 ) are not featured in the database explorer: refresh the dabase display by pressing [F5] or by clicking again on the selected button "Functional data".
) are not featured in the database explorer: refresh the dabase display by pressing [F5] or by clicking again on the selected button "Functional data".
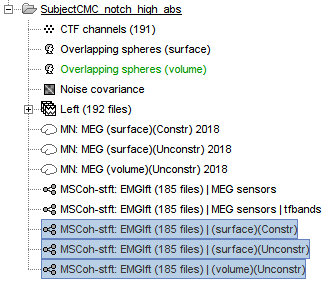
Surface
Double-click on the 1xN connectivity files for the two (surface) source spaces (constrained and unconstrained source orientations) to visualize the cortical maps. See main tutorial Display: Cortex surface for all available options. Pick the cortical location and frequency with the highest coherence value.
In the Surface tab: Smooth=30%, Amplitude=0%.
To compare visually different cortex maps, set manually the colormap range (e.g.[0 - 0.07])
Explore with coherence spectra with the frequency slider
The highest coherence value is located at 14.65 Hz, in the right primary motor cortex (precentral gyrus). To observe the coherence spectrum at a given location: right-click on the cortex > Source: Power spectrum.
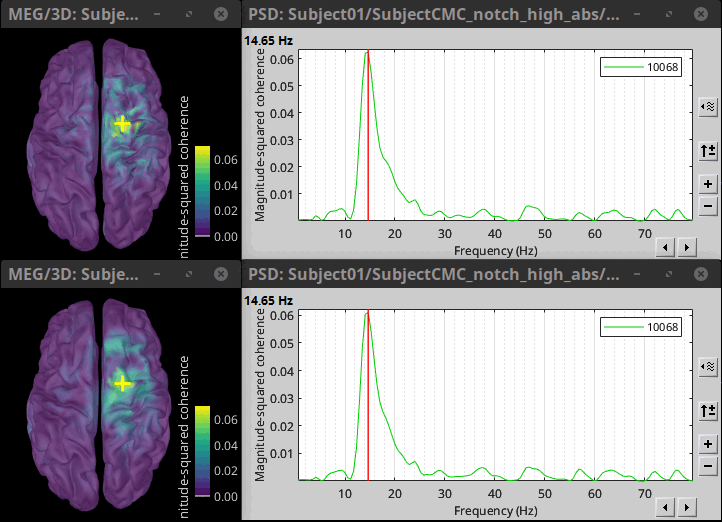
The constrained (top) and unconstrained (bottom) orientations coherence maps are qualitatively similar in terms of peak location and frequency. The unconstrained source map appears smoother because of the maximum aggregation across all three current directions at each brain location, as explained below. These maps results are similar to those obtained with the FieldTrip tutorial.
To obtain the 3D coordinates of the peak coherence location: right-click on the figure > Get coordinates. Then click on the right motor cortex with the crosshair cursor. Let's keep note of these coordinates for later comparision with the whole-head volume results below.
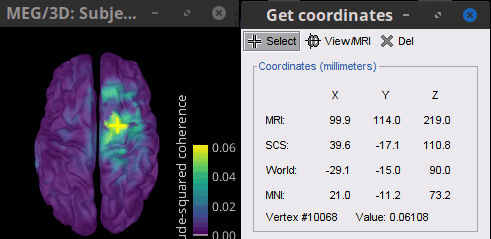
Volume
Double-click the 1xN connectivity file to display the (volume) source space.
- Set the frequency to 14.65 Hz, and the data transparency to 20% (Surface tab).
- Find the peak in the head volume by navigating in all 3 dimensions, or use the coordinates from the surface results as explained above.
Right-click on the figure > Anatomical atlas > None. The coherence value under the cursor is then shown on the top-right corner of the figure.
Method for constrained sources
For cortical and orientation-constrained sources, each vertex in the source grid is associated with 1 time series. As such, when coherence is computed with the EMG signal (also consisting of one time series), this result is 1 coherence spectrum per vertex. In other words, for each frequency bin, we obtain one coherence brain map.
Method for unconstrained sources
In the case of orientation-unconstrained (either surface or volume) sources, each vertex (source location) is associated with 3 time series, each one corresponding to the X, Y and Z directions. Thus, a dimension reduction across directions is needed. In this tutorial the dimension reduction consisted in first flattening the 3D maps to a single orientation with the PCA method. This results in a source model similar to that with orientation-constrained sources, thus, coherence is then computed as with orientation-constrained (described above).
PCA Options: If PCA is selected as the scout function or for flattening unconstrained maps, additional options are available through this Edit button.
Another alternative to address the issue of the dimension reduction across directions when estimating coherence with unconstrained sources is to compute coherence between the EMG signal (one time series) and each of the 3 time series (one for each source orientation: x, y and z) associated with each source location, resulting in 3 coherence spectra per source location. Then, these coherence spectra are collapsed into a single coherence spectrum by keeping the maximum coherence value across those in the three directions for each frequency bin. The choice of the maximum statistic is empirical: it is not rotation invariant (if the specifications of the NAS/LPA/RPA fiducials change, the x,y,z axes also change, which may influence connectivity measures quantitatively). Nevertheless, our empirical tests showed this option produced smooth and robust (to noise) connectivity estimates.
Below the comparison of the coherence between the EMG sensor and unconstrained surface sources using: PCA flattening in the source space (left) and "Flattening of the coherence, by maximum coherence across directions"
Coherence: EMG x Scouts
We have now obtained coherence spectra at each of the 15002 brain source locations. This is a very large amount of data to shift through. We therefore recommend to further reduce the dimensionality of the source space by using a surface- or volume-parcellation scheme. In Brainstorm vernacular, this can be achieved via an atlas consisting of scout regions. See the Scout Tutorial for detailed information about atlases and scouts in Brainstorm. To compute scout-wise coherence, it is necessary to specify two parameters to indicate how the data is aggregated in each scout:
1. The scout function (for instance, the mean), and
2. When the scout aggregation procedure if performed, either before or after the coherence computation).
Method for constrained sources
When the scout function is used with constrained sources, the computation of coherence is carried as follows:
If the scout function is applied before: The scout function (e.g., mean) is applied for all the source time series in the scout; resulting in 1 time series per scout. This scout time series is then used to compute coherence with the reference signal (here, with EMG) resulting in 1 coherence spectrum per scout.
If the scout function applied after: coherence is first computed between the reference signal (here, EMG) and each source time series in the scout, resulting in N coherence spectra, with N being the number of sources in the scout. Finally the scout function is applied to the resulting coherence spectra for each source within the scout to obtain one coherence spectrum per scout. This is a considerably more greedy option that the "before" option above, as it computes coherence measures between the EMG signal and each of the 15000 elementary source signals. This requires more computing resources; see introduction for an example.
Method for unconstrained sources
As ?described above, when computing coherence with unconstrained sources it is necessary to perform dimension reduction across directions. When this reduction is done by first flattening the unconstrained sources, is performed on unconstrained sources coherence computations is carried out as described in the ?previous section. However, if the sources are not flattened before computing coherence is carried out as follows:
If the scout function is applied before, the scout function (e.g., mean) is applied for each direction on the elementary source time series in the scout; resulting in 3 time series per scout, one time series per orientation (x, y, z). These scout time series are then used to compute coherence with the reference signal (here, with EMG), and the coherence spectra for each scout are aggregated across the x, y and z dimensions, with the max function as shown previously, to obtain 1 coherence spectrum per scout.
If scout function is applied after, coherence is computed between the reference signal (here, EMG) and each elementary sources (number of sources in each scout x 3 orientations). Then the coherence spectra for each elementary source are aggregated across the x, y and z dimensions. Finally the scout function is applied to the resulting coherence spectra for each elementary source within the scout. This is a considerably more greedy option that the "before" option above, as it computes coherence measures between the EMG signal and each of the 45000 elementary source signals. This requires more computing resources; see introduction for an example.
Example
Here, we will compute the coherence from scouts, using mean as scout function with the before option. We will use the Schaefer 100 parcellation atlas on to the orientation-constrained source map.
Use Search Database (
 ) to select the Left trials with their respective (surface)(Constr) source maps, as shown in the previous section.
) to select the Left trials with their respective (surface)(Constr) source maps, as shown in the previous section. In the Process2: Left trial group into both the Files A and Files B boxes. Select Process recordings (
 ) for Files A, and Process sources (
) for Files A, and Process sources ( ) for Files B.
) for Files B. 
Open the Pipeline editor:
Add the process Connectivity > Coherence AxB [2023] with the following parameters:
Time window = All file
Source channel (A) = EMGlft
Check Use scouts (B)
From the menu at the right, select Schaefer_100_17net
- Select all the scouts
Scout function: Apply: before
Scout function: mean
Select Magnitude squared coherence as Connectivity metric
Select Fourier transform as Time-freq decomposition method
Click in the [Edit] button to set the parameters of the Fourier transform:
Select Matlab's FFT defaults as Frequency definition
FT window length = 0.5 s
FT window overlap = 50%
Highest frequency of interest = 80 Hz
Time resolution = None
Estimate & save = across combined files/epochs
Add the process File > Add tag with the following parameters:
Tag to add = (surface)(Constr)
Select Add to file name
- Run the pipeline
|
|
|
Double-click on the new file: the coherence spectra are not displayed on the cortex; they are plotted for each scout. The 1xN connectivity file can also be shown as an image as displayed below:
|
|
|
Note that at 14.65 Hz, the two highest peaks correspond to the SomMotA_4 R and SomMotA_2 R scouts, both located over the right primary motor cortex.
The brain parcellation scheme is the user's decision. We recommend the Schaefer100 atlas used here by default.
Coherence Scouts x Scouts
When computing whole-brain connectomes (i.e., NxN connectivity matrices between all brain sources), additional technical considerations need to be taken into account: the number of time series is typically too large to be computationally tractable on most workstations (see introduction above); if using unconstrained source maps, with 3 time series at each brain location, this requires an extra step to derive connectivity estimates.
The last two sections explained the computation of coherence measures between one reference sensor (EMG) and brain source maps, with sensor x sources and sensor x scouts scenarios, both with constrained and unconstrained source orientation models.
This section explains dimension reduction (across number of sources) using scouts to compute scouts x scouts coherence, for constrained and unconstrained source maps. As in the ?sensor-scout coherence scenario, it is necessary to specify two parameters to indicate how the data is aggregated in each scout:
1. The scout function (for instance, the mean), and
2. When the scout aggregation procedure if performed, either before or after the coherence computation).
The sections below explain the processing steps for the cases of constrained and unconstrained sources, for coherence between two scouts (Scout-1 and Scout-2). In either case, the outcome is one coherence spectrum per pair of scouts.
Method for constrained sources
The computation steps depend of when the scout function is applied:
If the scout function is applied before: The scout function (e.g., mean) is applied for all the source time series in each scout; resulting in 1 time series per scout. These scout time series are then used to compute coherence resulting in 1 coherence spectrum per scout.
If the scout function applied after: coherence is first computed between each combination of souces-scout1 and sources-scout2 resulting in N coherence spectra, with N being the product of sources-scout1 and sources-scout2. Finally the scout function is applied twice (once for each scout) to the resulting coherence spectra to obtain one coherence spectrum per scout pair. This is a considerably more greedy option that the "before" option above, as it computes coherence measures each source in scout1 and each source in scout2. This requires more computing resources; see introduction for an example.
Method for unconstrained sources
As ?described above, when computing coherence with unconstrained sources it is necessary to perform dimension reduction across directions. When this reduction is done by first flattening the unconstrained sources, is performed on unconstrained sources coherence computations is carried out as described in the ?previous section. However, if the sources are not flattened before computing coherence is carried out as follows:
Before: The scout function (e.g., mean) is applied on each direction on the elementary source time series each scout, which results in 3 time series per elementary direction, per scout. These scouts time series are then used to compute coherence measures, yielding to 9 coherence spectra (1x-2x, 1x-2y, 1x-2z, 1y-2x, ..., 1z-2z) for each pair of scouts. The resulting coherence spectra are finally aggregated across the 9 dimensions with maximum per frequency bin, resulting in one single coherence spectrum for each pair of scouts.
After: With this option, coherence is first computed between each pair of source locations (each with 3 time series) in the two scouts, yielding to 9 coherence spectra for each pair of sources in the scouts. Then, these coherence spectra are aggregated across the 9 combinations of dimensions (1x-2x, 1x-2y, 1x-2z, 1y-2x, ..., 1z-2z) to obtain one single coherence spectrum for each pair of sources within the scouts. Finally, the scout function (e.g., mean) is applied twice (once per scout) to the coherence spectra for each source within the scouts to obtain one single coherence spectrum for each pair of scouts. Applying the scout function "after" is a considerably more greedy option that the applying the scout function "before", as it computes coherence measures between all 45000x45000 elementary source signals, and this number is multiplied by the number of frequency bins. This requires more computing resources; see the introduction for an example.![]()
Example
Here, we will compute the coherence from scouts, using mean as scout function with the before option. We will use the Schaefer 100 parcellation atlas on to the orientation-constrained source map.
Use Search Database (
 ) to select the Left trials with their respective (surface)(Constr) source maps, as shown in the previous section.
) to select the Left trials with their respective (surface)(Constr) source maps, as shown in the previous section. In the Process2: Left trial group into both the Files A and Files B boxes. Select Process recordings (
 ) for Files A, and Process sources (
) for Files A, and Process sources ( ) for Files B.
) for Files B. 
Open the Pipeline editor:
Add the process Connectivity > Coherence AxB [2023] with the following parameters:
Time window = All file
Source channel (A) = EMGlft
Check Use scouts (B)
From the menu at the right, select Schaefer_100_17net
- Select all the scouts
Scout function: Apply: before
Scout function: mean
Select Magnitude squared coherence as Connectivity metric
Select Fourier transform as Time-freq decomposition method
Click in the [Edit] button to set the parameters of the Fourier transform:
Select Matlab's FFT defaults as Frequency definition
FT window length = 0.5 s
FT window overlap = 50%
Highest frequency of interest = 80 Hz
Time resolution = None
Estimate & save = across combined files/epochs
Add the process File > Add tag with the following parameters:
Tag to add = (surface)(Constr)
Select Add to file name
- Run the pipeline
|
|
|
Double-click on the new file: the coherence spectra are not displayed on the cortex; they are plotted for each scout. The 1xN connectivity file can also be shown as an image as displayed below:
|
|
|
Note that at 14.65 Hz, the two highest peaks correspond to the SomMotA_4 R and SomMotA_2 R scouts, both located over the right primary motor cortex.
The brain parcellation scheme is the user's decision. We recommend the Schaefer100 atlas used here by default.
Method for mixed source models
Mixed models refer to source models with multiple regions that may include surface and volume regions, and regions with different source orientation constraints. To compute connectivity between surface and volume regions, you must first create volume scouts for the volume regions of the mixed model, and then use the process2 tab with the same files on both sides. On one side you can select surface scouts, and the other, the volume scouts you created. For connectivity between surface regions only, or volume regions only, use the process1 tab.
Additional documentation
Articles
Conway BA, Halliday DM, Farmer SF, Shahani U, Maas P, Weir AI, et al.
Synchronization between motor cortex and spinal motoneuronal pool during the performance of a maintained motor task in man.
The Journal of Physiology. 1995 Dec 15;489(3):917–24.Kilner JM, Baker SN, Salenius S, Hari R, Lemon RN.
Human Cortical Muscle Coherence Is Directly Related to Specific Motor Parameters.
J Neurosci. 2000 Dec 1;20(23):8838–45.Liu J, Sheng Y, Liu H.
Corticomuscular Coherence and Its Applications: A Review.
Front Hum Neurosci. 2019 Mar 20;13:100.Sadaghiani S, Brookes MJ, Baillet S.
Connectomics of human electrophysiology.
NeuroImage. 2022 Feb;247:118788.
Tutorials
Tutorial: Functional connectivity
Tutorial: Source estimation
Tutorial: Volume source estimation
Tutorial: Scouts
Tutorial: Connectivity graphs
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_coherence.m