|
Size: 2943
Comment:
|
Size: 16142
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Realistic head model: FEM with Duneuro = ''Authors: Takfarinas MEDANI,'' |
= Realistic head model: FEM with DUNEuro = ''Authors: [[https://neuroimage.usc.edu/brainstorm/AboutUs/tmedani|Takfarinas Medani]], Juan Garcia-Prieto, Francois Tadel, Sophie Schrader, Anand Joshi, Christian Engwer, Carsten Wolters, John Mosher and Richard Leahy '' |
| Line 4: | Line 4: |
| This forward model uses a finite element element method (FEM) and was developed by the '''...(l''''''ink)'''. It uses the volume mesh of the realistic head model. The goal of this forward solution is mostly for '''EEG users''', to provide more accurate results than the spherical models and more complex geometry than the BEM. It is not necessary for MEG users, as the "overlapping spheres" method gives similar results but much faster. This method is illustrated using the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG and epilepsy]] or similar data ('''todo'''). | {{attachment:logo_duneuro.png||align="right",height="82",width="187"}} |
| Line 6: | Line 6: |
| refe to these point : http://duneuro.org/ | This tutorial explains how to use [[https://www.medizin.uni-muenster.de/duneuro/startseite.html|DUNEuro]] to compute the forward model using the '''finite element method''' ('''FEM'''). The FEM methods use a realistic volume mesh of the head generated from the segmentation of the MRI. The FEM models provide more accurate results than the spherical forward models and more realistic geometry and tissue properties than the [[Tutorials/TutBem|BEM]] methods. Its applications include accurate source localization in MEG/EEG/sEEG/ECoG and TMS/TDCS optimizations. |
| Line 8: | Line 8: |
| and to the documentation from here : | The scope of this page is limited to a '''basic example''' (head model with 3 layers). More advanced options for mesh generation and forward model computation are discussed in other tutorials: [[Tutorials/FemMesh|FEM mesh generation]], [[Tutorials/FemTensors|FEM tensors estimation]], [[Tutorials/FemMedianNerveCharm|FEM median nerve example]]. We assume that you have already followed the [[Tutorials|introduction tutorials]], we will not discuss the general principles of forward modeling here. |
| Line 10: | Line 10: |
| https://gitlab.dune-project.org/duneuro/duneuro/wikis/home | <<TableOfContents(2,2)>> |
| Line 12: | Line 12: |
| Here we will describe the FEM and run some examples | == DUNEuro == '''[[https://www.medizin.uni-muenster.de/duneuro/startseite.html|DUNEuro]] '''is an open-source C++ software library for solving partial differential equations (PDE) in neurosciences using mesh-based methods. It is based on the''' [[https://www.dune-project.org/|DUNE library]] '''and its main features include solving the EEG and MEG forward problem and providing simulations for brain stimulation. |
| Line 14: | Line 15: |
| == Duneuro == duneuro is an open-source C++ software library for solving partial differential equations in neurosciences using mesh bases methods. It is based on the DUNE library and its main features include solving the electroencephalography (EEG) and magnetoencephalography (MEG) forward problem and providing simulations for brain stimulation. |
As distributed on the [[http://gitlab.dune-project.org/duneuro/duneuro|DUNEuro GitLab]], the source code works only on Linux operating systems. Interfaces to Matlab and Python are possible, but you need to install and compile DUNEuro by yourself. For Brainstorm, we adapted this code and were able to generate '''binaries''' for the main operating systems''' '''(Windows, '''Linux '''and MacOS), which are '''downloaded automatically''' when needed as a [[Tutorials/Plugins|Brainstorm plugin]]. This project is available on our [[https://github.com/brainstorm-tools/bst-duneuro|GitHub repository bst-duneuro]]. |
| Line 17: | Line 17: |
| http://duneuro.org/ | We would like to thank the''' DUNEuro team '''for their help with this integration work: Carsten Wolters, Christian Engwer, Sophie Schrader, Andreas Nuessing, Yvonne Buschermöhle, Tim Erdbruegger, Marios Antonakakis, Johannes Vorwerk & Maria Carla Piastra. ''' {{attachment:duneuroFromDune.JPG||height="187",width="546"}} ''' If you use Brainstorm-DUNEuro please '''cite the following papers''' in your work: * Schrader, S., Westhoff, A., Piastra, M.C., Miinalainen, T., Pursiainen, S.,Vorwerk, J., Brinck, H., Wolters, C.H., Engwer, C., DUNEuro- A software toolbox for forward modeling in bioelectromagnetism, PLoS ONE, 16(6):e0252431 (2021). doi: [[https://urldefense.com/v3/__https://doi.org/10.1371/journal.pone.0252431__;!!LIr3w8kk_Xxm!-bMXySp_3t41y5-tXZHEiMKGFE32SAMz93NulthzqfHf4uA7QstTC15r64oFpQ$|https://doi.org/10.1371/journal.pone.0252431]] * Medani, T., Garcia-Prieto, J., Tadel, F., Schrader, S. Antonakakis, M., Joshi, A., Engwer, C., Wolters, C.H., Mosher, J., Leahy, R., "Realistic head modeling of electromagnetic brain activity: an integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions," Proc. SPIE 11595, Medical Imaging 2021: Physics of Medical Imaging, 1159554 (15 February 2021);https://doi.org/10.1117/12.2580935 If you want to learn more about DUNEuro, please visit to the following web pages: * [[https://www.medizin.uni-muenster.de/duneuro/forschung.html|DUNEuro main website]] * [[https://www.medizin.uni-muenster.de/duneuro/forschung.html|DUNEuro publications]] |
| Line 20: | Line 32: |
| Duneuro is build on top of DUNE Library, at this time (sept 2019) these libraries work only on Linux systems. However we are able to run generate binaries for windows and can be easily used from Matlab for all platforms. | In order to reproduce the computation present below on your computer, you need to fulfill all the conditions listed below. Alternatively, you can read this page as a reference documentation about DUNEuro and not try to reproduce the results. |
| Line 22: | Line 34: |
| However if you want to use the source code of Duneuro you can visite: http://duneuro.org/ | * You have a working copy of Brainstorm installed on your computer. * You have already followed the [[https://neuroimage.usc.edu/brainstorm/Introduction|introduction tutorials]], at least until #23. * You have followed the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG and Epilepsy]], as it is used for illustrating the computation. * '''DUNEuro''': Software installed automatically as a [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|Brainstorm plugin]]. * '''Iso2mesh''': Software installed automatically as a [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|Brainstorm plugin]]. |
| Line 24: | Line 40: |
| We also put here : | == FEM mesh == In order to use the FEM computations of the electromagnetic field (EEG/MEG), the volume mesh of the head is required. Brainstorm integrates most of the modern open-source tools to generate realistic head mesh, either from nested surface mesh or from individual MR images (T1 or T1 and T2). This tutorial describes only a simple approach based on three nested surfaces meshed in 3D with Iso2mesh. For the full list of available methods and description of options, please refer to the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh|FEM mesh generation]].''' ''' |
| Line 26: | Line 43: |
| The main steps you need in order to compile for windows are listed here : | * Select the protocol TutorialEpilepsy, created while following the tutorial [[Tutorials/Epilepsy|EEG and epilepsy]]. * Go to the anatomy view. * Select the three [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#BEM_surfaces|BEM surfaces]]: scalp, outer skull, inner skull. * Right-click > '''Generate FEM mesh''' > Iso2mesh-2021 > MergeMesh > Default options. <<BR>><<BR>> {{attachment:femMesh1.gif}} <<BR>> {{attachment:iso2mesh.gif}} ''' ''' * The FEM mesh appears in the database explorer after a short while. The first number indicates the number of vertices (i.e. nodes) of the tetrahedral mesh. To get the number of 3D elements (i.e. tetrahedrons) in this geometric model of the head: right-click on the file > File > View file contents. The structure of the file is describe in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#On_the_hard_drive|FEM mesh generation]].<<BR>><<BR>> {{attachment:femMesh2.gif}} * Double-click on the FEM file to display it. From the Surface tab, you can change the resection locations by moving the bottom sliders, and you can control the display of each of the three layers individually. Click on the layer button in the toolbar, then adjust the color and transparency of the corresponding mesh. The figure below represents the FEM mesh in the center and the initial BEM layers on the right. <<BR>><<BR>> {{attachment:femMesh3.gif}} |
| Line 28: | Line 50: |
| https://github.com/svdecomposer/brainstorm-duneuro | == FEM forward model == The '''forward model''' (or '''head model''' in the Brainstorm documentation and interface) describes how the electric activity in the '''source space''' (the cortex surface or a regular grid of volume points) influences the electric potential (EEG) or magnetic fields (MEG) at the level of the '''sensors'''. The FEM method uses the '''tetrahedral mesh''' computed above to establish this relationship. ''' ''' |
| Line 30: | Line 53: |
| We distribute the binary files for Windows/Ubunto 64bit systems, you don't need to installan extra Microsoft software package to run '''Duneuro''' | * Go the Anatomy view. Select the default FEM mesh and cortex surface you'd like to use for the computation (in case there is more than one in each category). The selected elements appear in green, double-click or right-click > Set as default to change the selection. * Go to the Functional view. Right-click on the channel file > '''Compute head model'''. <<BR>><<BR>> {{attachment:femCompute1.gif}} * Select '''MRI volume''', EEG: '''DUNEuro FEM''', select all the layers and the default conductivity values (scalp = inside the head surface, skull = inside the outer skull surface, brain = inside the inner skull surface).<<BR>><<BR>> {{attachment:femCompute2.gif}} * Volume source grid: '''Regular grid''', brain, 5mm. {{attachment:femCompute3.gif}} * Start the computation. Depending on the complexity of the problem (number of FEM elements, number of layers, number of source points, number of sensors and computation options), this can take anywhere between a few minutes to a few days. In this example, if you selected the correct files and options, it should only take a few minutes. You may be prompted to download or update the bst-duneuro [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|plugin]]. <<BR>><<BR>> {{attachment:femCompute4.gif}} * When it's finished, you have obtain a new head model, which can be used for source estimation: see the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Source_analysis:_Volume|epilepsy tutorial]], the [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource|volume source estimation]] tutorial, or the [[https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation|introduction tutorial]]. |
| Line 32: | Line 60: |
| . Microsoft Visual C++ 2010 Redistributable Package (x64). | <<TAG(Advanced)>> |
| Line 34: | Line 62: |
| == FEM surfaces / Volume generation == === Volume generation from surface files === In this part you can generate your FEM mesh from surfaces that you can get fron the segmentation software (brainSuite, FreeSurfer ....). |
== DUNEuro options: Basic == When assuming '''isotropic conductivities''' for all the tissues, the DUNEuro basic options are limited to the selection of the tissues and their conductivities. |
| Line 38: | Line 65: |
| This process will | . {{attachment:duneuroBasic.gif}} {{attachment:duneuroTensors.gif}} |
| Line 40: | Line 67: |
| - merge the surfaces, | * '''FEM layers: '''Brainstorm reads the lists of tissues from the selected FEM mesh. Users can select the desired layers to include in the FEM computation. According to the modalities, the recommended selections are: * '''EEG''': Select all the layers * '''MEG''': Select only the inner layers (inside the inner skull = brain = white+gray+csf) * '''SEEG''': Select only the inner layers * '''ECOG''': Select only the inner layers * Any combination of modalities that includes MEG: select all the layers |
| Line 42: | Line 74: |
| - check the self intersecting | * '''FEM conductivities (isotropic):''' Left figure - Brainstorm proposes default conductivity values for each layer (derived from the SimBio software, see [[https://www.mrt.uni-jena.de/simbio/images/VorwerkChoRamppHamerKnoescheWolters_NeuroImage_2014_Webversion.pdf|this article]]). However, you have the possibility to change these values according to your model. |
| Line 44: | Line 76: |
| - fixe the size of the mesh | * '''FEM conductivities (anisotropic)''': Right figure - Advanced users can use information from '''diffusion imaging''' exams to estimate anisotropic '''conductivity tensors''' for each element of the FEM mesh. For more information, refer to the tutorial: [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemTensors|FEM tensors estimation]]. To remove previously computed tensors and restore the default isotropic approach, right-click on the FEM mesh > Clear FEM tensors. <<BR>><<BR>> {{attachment:clearFemTensors.JPG||height="300",width="220"}} |
| Line 46: | Line 78: |
| - generate the volum mesh | <<TAG(Advanced)>> |
| Line 48: | Line 80: |
| - visual checking ... | == DUNEuro options: Advanced [TODO] == In the DUNEuro options panel, click on the button '''Show details''' to access the advanced options. |
| Line 50: | Line 83: |
| - TODO : may be we can add some mesh auqlity measures ?? | . {{attachment:duneuroAdvanced.gif}} |
| Line 52: | Line 85: |
| * '''FEM layers & conductivities''': Discussed in the previous section. | |
| Line 53: | Line 87: |
| * '''FEM solver type''': (only the fitted FEM are integrated in Brainstorm at the moment) * '''CG''': Continuous Galerkin: the standard Lagrangian method. |
|
| Line 54: | Line 90: |
| === Volume generation from T1 MRI data === | * '''DC''': Discontinuous Galerkin: method that deals with some problems that appear occasionaly during meshing of very thin volumes. DC enforces a realistic modeling of the volume currents to attempt to mitigate these problems. |
| Line 56: | Line 92: |
| === Head model based on the level set approach === | * '''FEM source model''': * '''Venant''': The details are docuemented in this thesis: * Number of moments: * Reference length: * Weighting exponent: * Relaxation factor exponent: * Mixed moment: * Restrict: * '''Subtraction''': * intorderadd: * intorderadd_lb: * '''Partial integration''': * '''Source space''': * '''Shrink source space''': The dipoles are moved inward by the specified distance (in mm). * '''Force source space''': This is required when the dipoles are not within the gray matter. * '''Output options''': * '''Save transfer matrix''': save the transfer matrix within the tmp folder. This is usfeul if the user want to test different source model (PI, St-Venant) |
| Line 58: | Line 111: |
| == Forward model == === Head model === Number of layers, conductivity value, |
For more information about these methods, users can check [[https://www.sci.utah.edu/~wolters/PaperWolters/2016/Vorwerk_Dissertation_2016.pdf|Johannes Vorwerk's PhD thesis]]. |
| Line 62: | Line 113: |
| === Electrode model === Check the position of the electrodes and align to the head model (projection if needed) |
<<TAG(Advanced)>> |
| Line 65: | Line 115: |
| === Source model === | == Troubleshooting == DUNEuro binaries may crash for various reasons. We will try to describe the most frequent problems, however, in general terms, FEM forward modeling problems can be related with memory overload or extremely long computation times. For this reason it is always a good idea to reduce the size of the problem and test simpler solutions first, before attempting a fine grained computation. |
| Line 67: | Line 118: |
| === Advanced paramaters === | If you cannot find a solution, please post the full error message on the Brainstorm user forum (you can copy-paste the error message from the Matlab command window after closing the error message box). |
| Line 69: | Line 120: |
| === Grid error messages === * Error message: Coordinate is outside of the grid, or grid is not convex. * Explanation: Some dipoles are probably outside of the cortex, users need to correct the source space. * Try redefining the source model by making sure that the sources are not located out of the cortical volume. === Remove the neck === One way to reduce the size of the forward problem is to decrease the number of FEM elements in the head model. When the field of the MRI is large, you may have the mesh of the neck and even the shoulders. In most cases, it is safe to remove the lower part of the FEM mesh, below the nose and the brainstem. Right-click on the FEM mesh > '''Resect Neck'''. {{attachment:resectNeck.JPG||height="150",width="600"}} Once the process is finished, a new FEM mesh appears in the database, with a tag "resect". The following figure shows the model before (743828 vertices / 4079587 elements) and after resection (613955 vertices / 3400957 elements). It will reduce the size of the problem by 20%. {{attachment:FemMeshAllandResect.JPG||height="150",width="600"}} === Linux Operating Systems === Some users have reported the following error when computing the head model with DUNEuro in Linux OS. {{{ Error while loading shared libraries: libldl.so.2: cannot open shared object file: No such file or directory }}} The problem is that DUNEuro requires some libraries in order to work properly. These dependencies should be in a folder where they can be located. The error message occurs when this dependencies eithera are missing in your system or in your path. In order to solve it, you can try to find any library with the same name. Be careful, ''libdl.so'' and ''libldl.so'' are not the same thing. {{{ find -name "libldl*" /usr }}} If you find a ''libldl'' library, but it is not in the correct path, try copying the library (or a link to it) to ''/usr/lib''. If you cannot find any related library in your system, try installing the following packages to force that dependency being installed in your system. For instance, in Ubuntu you can run: {{{ sudo apt install libldl2 libspar2 libdune-grid-dev libdune-istl-dev }}} |
|
| Line 70: | Line 153: |
| == Reported Errors & alternative solution == '''<<TAG(Advanced)>>''' |
==== Related tutorials ==== * [[Tutorials/FemMesh|FEM mesh generation]] * [[Tutorials/FemTensors|FEM tensors estimation]] * [[Tutorials/FemMedianNerveCharm|FEM median nerve example]] |
| Line 73: | Line 158: |
| == The MEEG forward problem with the FEM == == License == == Reference == |
==== DUNEuro references ==== * DUNEuro wiki: https://gitlab.dune-project.org/duneuro/duneuro/wikis/home * DUNEuro website: https://www.medizin.uni-muenster.de/duneuro/startseite.html * List of the parameters: https://docs.google.com/spreadsheets/d/1MqURQsszn8Qj3-XRX_Z8qFFnz6Yl2-uYALkV-8pJVaM/edit#gid=0 ==== Brainstorm-DUNEuro integration ==== * https://github.com/brainstorm-tools/bst-duneuro/issues/1 * https://github.com/brainstorm-tools/brainstorm3/issues/242 * https://github.com/brainstorm-tools/brainstorm3/issues/185 '''Brainstorm user forum''' * Sources in white matter: https://neuroimage.usc.edu/forums/t/each-source-is-in-which-layer-of-the-mesh/31887/10 |
Realistic head model: FEM with DUNEuro
Authors: Takfarinas Medani, Juan Garcia-Prieto, Francois Tadel, Sophie Schrader, Anand Joshi, Christian Engwer, Carsten Wolters, John Mosher and Richard Leahy

This tutorial explains how to use DUNEuro to compute the forward model using the finite element method (FEM). The FEM methods use a realistic volume mesh of the head generated from the segmentation of the MRI. The FEM models provide more accurate results than the spherical forward models and more realistic geometry and tissue properties than the BEM methods. Its applications include accurate source localization in MEG/EEG/sEEG/ECoG and TMS/TDCS optimizations.
The scope of this page is limited to a basic example (head model with 3 layers). More advanced options for mesh generation and forward model computation are discussed in other tutorials: FEM mesh generation, FEM tensors estimation, FEM median nerve example. We assume that you have already followed the introduction tutorials, we will not discuss the general principles of forward modeling here.
Contents
DUNEuro
DUNEuro is an open-source C++ software library for solving partial differential equations (PDE) in neurosciences using mesh-based methods. It is based on the DUNE library and its main features include solving the EEG and MEG forward problem and providing simulations for brain stimulation.
As distributed on the DUNEuro GitLab, the source code works only on Linux operating systems. Interfaces to Matlab and Python are possible, but you need to install and compile DUNEuro by yourself. For Brainstorm, we adapted this code and were able to generate binaries for the main operating systems (Windows, Linux and MacOS), which are downloaded automatically when needed as a Brainstorm plugin. This project is available on our GitHub repository bst-duneuro.
We would like to thank the DUNEuro team for their help with this integration work: Carsten Wolters, Christian Engwer, Sophie Schrader, Andreas Nuessing, Yvonne Buschermöhle, Tim Erdbruegger, Marios Antonakakis, Johannes Vorwerk & Maria Carla Piastra.
If you use Brainstorm-DUNEuro please cite the following papers in your work:
Schrader, S., Westhoff, A., Piastra, M.C., Miinalainen, T., Pursiainen, S.,Vorwerk, J., Brinck, H., Wolters, C.H., Engwer, C., DUNEuro- A software toolbox for forward modeling in bioelectromagnetism, PLoS ONE, 16(6):e0252431 (2021). doi: https://doi.org/10.1371/journal.pone.0252431
Medani, T., Garcia-Prieto, J., Tadel, F., Schrader, S. Antonakakis, M., Joshi, A., Engwer, C., Wolters, C.H., Mosher, J., Leahy, R., "Realistic head modeling of electromagnetic brain activity: an integrated Brainstorm-DUNEuro pipeline from MRI data to the FEM solutions," Proc. SPIE 11595, Medical Imaging 2021: Physics of Medical Imaging, 1159554 (15 February 2021);https://doi.org/10.1117/12.2580935
If you want to learn more about DUNEuro, please visit to the following web pages:
Requirements
In order to reproduce the computation present below on your computer, you need to fulfill all the conditions listed below. Alternatively, you can read this page as a reference documentation about DUNEuro and not try to reproduce the results.
- You have a working copy of Brainstorm installed on your computer.
You have already followed the introduction tutorials, at least until #23.
You have followed the tutorial EEG and Epilepsy, as it is used for illustrating the computation.
DUNEuro: Software installed automatically as a Brainstorm plugin.
Iso2mesh: Software installed automatically as a Brainstorm plugin.
FEM mesh
In order to use the FEM computations of the electromagnetic field (EEG/MEG), the volume mesh of the head is required. Brainstorm integrates most of the modern open-source tools to generate realistic head mesh, either from nested surface mesh or from individual MR images (T1 or T1 and T2). This tutorial describes only a simple approach based on three nested surfaces meshed in 3D with Iso2mesh. For the full list of available methods and description of options, please refer to the tutorial FEM mesh generation.
Select the protocol TutorialEpilepsy, created while following the tutorial EEG and epilepsy.
- Go to the anatomy view.
Select the three BEM surfaces: scalp, outer skull, inner skull.
Right-click > Generate FEM mesh > Iso2mesh-2021 > MergeMesh > Default options.
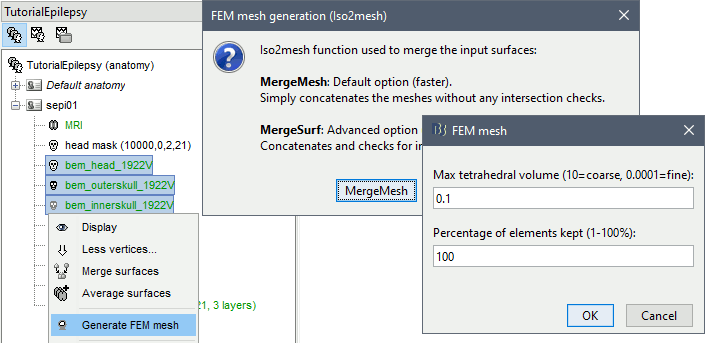
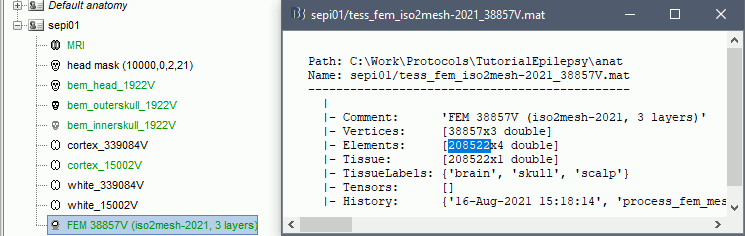
The FEM mesh appears in the database explorer after a short while. The first number indicates the number of vertices (i.e. nodes) of the tetrahedral mesh. To get the number of 3D elements (i.e. tetrahedrons) in this geometric model of the head: right-click on the file > File > View file contents. The structure of the file is describe in the tutorial FEM mesh generation.
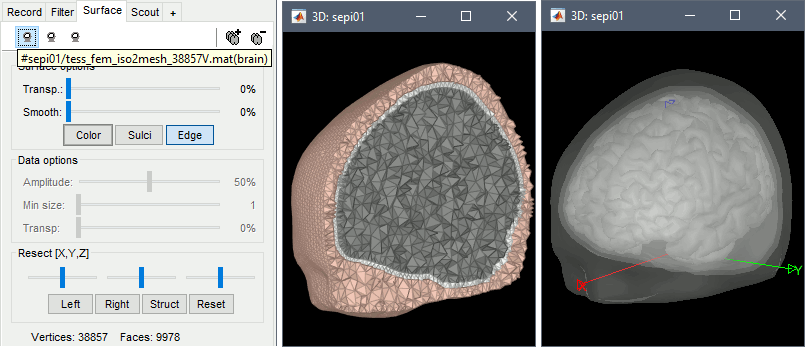
Double-click on the FEM file to display it. From the Surface tab, you can change the resection locations by moving the bottom sliders, and you can control the display of each of the three layers individually. Click on the layer button in the toolbar, then adjust the color and transparency of the corresponding mesh. The figure below represents the FEM mesh in the center and the initial BEM layers on the right.
FEM forward model
The forward model (or head model in the Brainstorm documentation and interface) describes how the electric activity in the source space (the cortex surface or a regular grid of volume points) influences the electric potential (EEG) or magnetic fields (MEG) at the level of the sensors. The FEM method uses the tetrahedral mesh computed above to establish this relationship.
Go the Anatomy view. Select the default FEM mesh and cortex surface you'd like to use for the computation (in case there is more than one in each category). The selected elements appear in green, double-click or right-click > Set as default to change the selection.
Go to the Functional view. Right-click on the channel file > Compute head model.
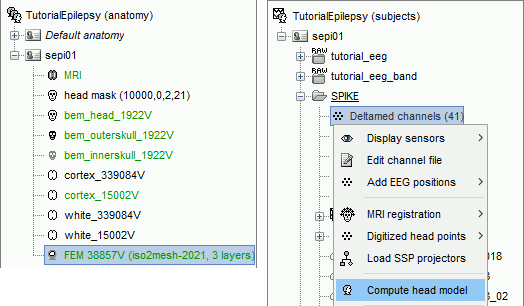
Select MRI volume, EEG: DUNEuro FEM, select all the layers and the default conductivity values (scalp = inside the head surface, skull = inside the outer skull surface, brain = inside the inner skull surface).
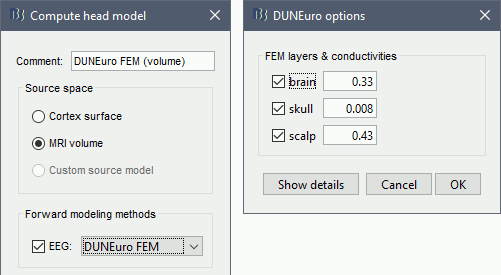
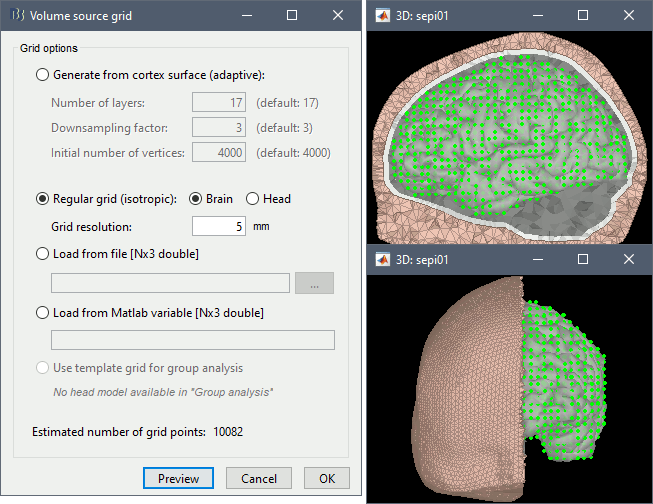
Volume source grid: Regular grid, brain, 5mm.
Start the computation. Depending on the complexity of the problem (number of FEM elements, number of layers, number of source points, number of sensors and computation options), this can take anywhere between a few minutes to a few days. In this example, if you selected the correct files and options, it should only take a few minutes. You may be prompted to download or update the bst-duneuro plugin.
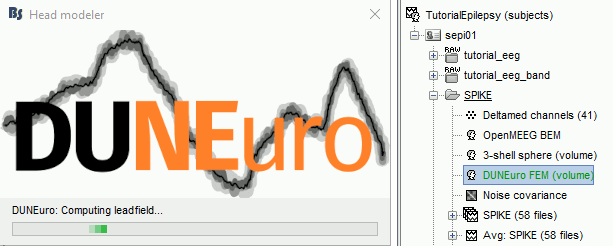
When it's finished, you have obtain a new head model, which can be used for source estimation: see the epilepsy tutorial, the volume source estimation tutorial, or the introduction tutorial.
DUNEuro options: Basic
When assuming isotropic conductivities for all the tissues, the DUNEuro basic options are limited to the selection of the tissues and their conductivities.
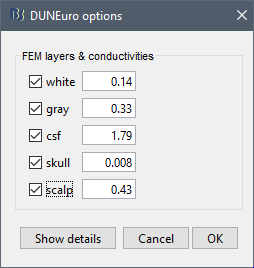
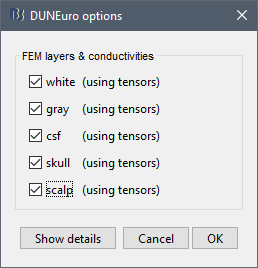
FEM layers: Brainstorm reads the lists of tissues from the selected FEM mesh. Users can select the desired layers to include in the FEM computation. According to the modalities, the recommended selections are:
EEG: Select all the layers
MEG: Select only the inner layers (inside the inner skull = brain = white+gray+csf)
SEEG: Select only the inner layers
ECOG: Select only the inner layers
- Any combination of modalities that includes MEG: select all the layers
FEM conductivities (isotropic): Left figure - Brainstorm proposes default conductivity values for each layer (derived from the SimBio software, see this article). However, you have the possibility to change these values according to your model.
FEM conductivities (anisotropic): Right figure - Advanced users can use information from diffusion imaging exams to estimate anisotropic conductivity tensors for each element of the FEM mesh. For more information, refer to the tutorial: FEM tensors estimation. To remove previously computed tensors and restore the default isotropic approach, right-click on the FEM mesh > Clear FEM tensors.
DUNEuro options: Advanced [TODO]
In the DUNEuro options panel, click on the button Show details to access the advanced options.
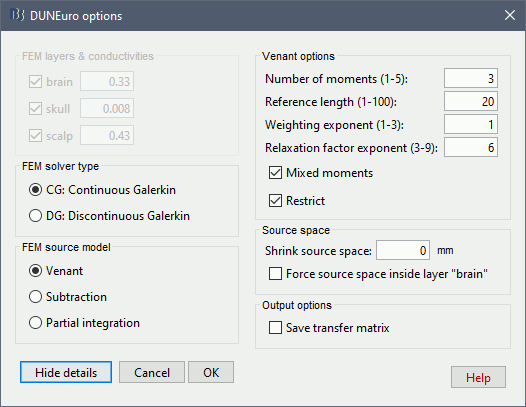
FEM layers & conductivities: Discussed in the previous section.
FEM solver type: (only the fitted FEM are integrated in Brainstorm at the moment)
CG: Continuous Galerkin: the standard Lagrangian method.
DC: Discontinuous Galerkin: method that deals with some problems that appear occasionaly during meshing of very thin volumes. DC enforces a realistic modeling of the volume currents to attempt to mitigate these problems.
FEM source model:
Venant:
The details are docuemented in this thesis:
- Number of moments:
- Reference length:
- Weighting exponent:
- Relaxation factor exponent:
- Mixed moment:
- Restrict:
Subtraction:
- intorderadd:
- intorderadd_lb:
Partial integration:
Source space:
Shrink source space: The dipoles are moved inward by the specified distance (in mm).
Force source space: This is required when the dipoles are not within the gray matter.
Output options:
Save transfer matrix: save the transfer matrix within the tmp folder. This is usfeul if the user want to test different source model (PI, St-Venant)
For more information about these methods, users can check Johannes Vorwerk's PhD thesis.
Troubleshooting
DUNEuro binaries may crash for various reasons. We will try to describe the most frequent problems, however, in general terms, FEM forward modeling problems can be related with memory overload or extremely long computation times. For this reason it is always a good idea to reduce the size of the problem and test simpler solutions first, before attempting a fine grained computation.
If you cannot find a solution, please post the full error message on the Brainstorm user forum (you can copy-paste the error message from the Matlab command window after closing the error message box).
Grid error messages
- Error message: Coordinate is outside of the grid, or grid is not convex.
- Explanation: Some dipoles are probably outside of the cortex, users need to correct the source space.
- Try redefining the source model by making sure that the sources are not located out of the cortical volume.
Remove the neck
One way to reduce the size of the forward problem is to decrease the number of FEM elements in the head model. When the field of the MRI is large, you may have the mesh of the neck and even the shoulders. In most cases, it is safe to remove the lower part of the FEM mesh, below the nose and the brainstem. Right-click on the FEM mesh > Resect Neck.
Once the process is finished, a new FEM mesh appears in the database, with a tag "resect". The following figure shows the model before (743828 vertices / 4079587 elements) and after resection (613955 vertices / 3400957 elements). It will reduce the size of the problem by 20%.
Linux Operating Systems
Some users have reported the following error when computing the head model with DUNEuro in Linux OS.
Error while loading shared libraries: libldl.so.2: cannot open shared object file: No such file or directory
The problem is that DUNEuro requires some libraries in order to work properly. These dependencies should be in a folder where they can be located. The error message occurs when this dependencies eithera are missing in your system or in your path. In order to solve it, you can try to find any library with the same name. Be careful, libdl.so and libldl.so are not the same thing.
find -name "libldl*" /usr
If you find a libldl library, but it is not in the correct path, try copying the library (or a link to it) to /usr/lib.
If you cannot find any related library in your system, try installing the following packages to force that dependency being installed in your system. For instance, in Ubuntu you can run:
sudo apt install libldl2 libspar2 libdune-grid-dev libdune-istl-dev
Additional documentation
Related tutorials
DUNEuro references
DUNEuro wiki: https://gitlab.dune-project.org/duneuro/duneuro/wikis/home
DUNEuro website: https://www.medizin.uni-muenster.de/duneuro/startseite.html
List of the parameters: https://docs.google.com/spreadsheets/d/1MqURQsszn8Qj3-XRX_Z8qFFnz6Yl2-uYALkV-8pJVaM/edit#gid=0
Brainstorm-DUNEuro integration
Brainstorm user forum
Sources in white matter: https://neuroimage.usc.edu/forums/t/each-source-is-in-which-layer-of-the-mesh/31887/10
