|
Size: 13333
Comment:
|
Size: 18949
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Realistic head model: FEM mesh generation = '''[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]''' |
= FEM mesh generation = ''Authors: [[https://neuroimage.usc.edu/brainstorm/AboutUs/tmedani#preview|Takfarinas Medani]], Francois Tadel'' |
| Line 4: | Line 4: |
| ''Authors: Takfarinas Medani '' | FEM forward modeling requires the construction of a 3D model of thee head tissues. The volume of the head is divided in small geometrical elements with 4 faces (tetrahedrons) or 6 faces (hexahedrons). Each element is associated with a type of biological tissue (e.g. white matter, gray matter, CSF, skull, skin) and electrical conductivity properties. |
| Line 6: | Line 6: |
| <<TableOfContents(2,2)>> | This page lists the methods integrated with Brainstorm to generate 3D meshes of the head. For a generic introduction to FEM in Brainstorm, refer to the tutorials: [[https://neuroimage.usc.edu/brainstorm/Tutorials/Duneuro|Realistic head model: FEM with DUNEuro]] and [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMedianNerveCharm|FEM median nerve example]]. |
| Line 8: | Line 8: |
| == Introduction == This tutorial presents the methods integrated to brainstorm used to generate the FEM mesh. |
<<TableOfContents(3,2)>> |
| Line 11: | Line 10: |
| The FEM mesh is required for the finite element method computation. The FEM computation could be used for most of the known modalities: EEG/MEG forward problem, TMS or TDSC stimulation and for intracranial modalities like sEEG and ECOG. | == Generate FEM mesh == FEM meshes can be computed from surfaces (as the ones generated for the [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#BEM_surfaces|BEM models]]) or from MRI volumes (T1w and/or T2w). The methods that are available when using the popup menu '''Generate FEM mesh''' depend on the selected inputs. |
| Line 13: | Line 13: |
| In this tutorial, we present the different methods available with brainstorm to generate the FEM mesh and how to use them from the brainstorm GUI. | === Surfaces === Select a list of surfaces representing the separation between different tissues (holding the CTRL or SHIFT key), then right-click on any of them. The software [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#Iso2mesh|Iso2mesh]] can create a tetrahedral mesh to represent the tissues between these different layers. |
| Line 15: | Line 16: |
| == Mesh tools == Brainstorm integrates a list of open-source tools. These tools are commonly used by the FEM community to generate either tetrahedral or hexahedra mesh. |
{{attachment:callSurf.gif}} |
| Line 18: | Line 18: |
| Here is the list of the available methods in brainstorm: | === T1 MRI === Right-click on a T1 MRI available in the database. Typically, this is the default MRI volume displayed in green in the subject folder. Methods available: [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#Brain2mesh|Brain2mesh]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#SimNIBS_3:_headreco|SimNIBS3]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#SimNIBS_4:_charm|SimNIBS4]], [[http://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#ROAST|ROAST]], [[http://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#FieldTrip|FieldTrip]]. |
| Line 20: | Line 21: |
| - [[http://iso2mesh.sourceforge.net/cgi-bin/index.cgi?Doc/FunctionList|iso2mesh]]: this option merges the brainstorm surfaces available on the subject and then generate the tetrahedral mesh. | {{attachment:callT1.gif}} |
| Line 22: | Line 23: |
| - [[http://mcx.space/brain2mesh/|Brain2mesh]]: this option uses the MRI available on the subject, then it calls [[https://www.fil.ion.ucl.ac.uk/spm/software/spm12/|SPM]]segmentation of the volume into 5 tissues (white, gray, SCF, skull and skin). After that, it calls iso2mesh (internally) to generate the tetrahedral mesh. | === T1+T2 MRI === Select the T1+T2 volumes, then right-click on any of them. The different files are identified based on the tags "T1" and "T2" the file names (as displayed in the Brainstorm database explorer). If these identification tags are not found in the file names, the default MRI (in green) is used as the T1, the other as the T2. If none of the is the default MRI, the first selected file is used as the T1, the second is used as the T2. Methods available: [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#Brain2mesh|Brain2mesh]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#SimNIBS_3:_headreco|SimNIBS3]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#SimNIBS_4:_charm|SimNIBS4]], [[http://neuroimage.usc.edu/brainstorm/Tutorials/FemMesh#ROAST|ROAST]]. |
| Line 24: | Line 26: |
| - [[https://simnibs.github.io/simnibs/build/html/index.html|SimNibs]] : this option, recommended for a realistic model, calls the [[https://simnibs.github.io/simnibs/build/html/documentation/command_line/headreco.html|headreco]]process, it uses the MRIs of the subject, and then calls SPM and [[http://www.neuro.uni-jena.de/cat/|CAT]] for the segmentation. Then the mesh generation is performed internally by integrated tools (netgen, gmesh and meshfixe). | {{attachment:callT1T2.gif}} |
| Line 26: | Line 28: |
| - [[http://www.fieldtriptoolbox.org/|Fieldtrip]] : this option call the Fieldtrip pipeline, based on the segmentation of the MRI then the hexahedral mesh generation. | === Anatomy folder === If you right-click on the subject folder > Generate FEM mesh, then Brainstorm offers all the possible options, even the ones that are not applicable to this specific subject. |
| Line 28: | Line 31: |
| You can display the full list and a short description by right click on the MRI of the subject and then click the item "Generate FEM Mesh" | * '''Volume ''''''method''': If using a method based on MRI volumes, the T1 and T2 volumes are detected among all the volumes available based on the tags "T1" and "T2" in the file names (make sure only one file as each of these tags), otherwise use only the default MRI (in green) as the T1. * '''Surface method''': If using a method based on surfaces, the default surfaces (in green) from three categories as selected: the inner skull, the outer skull and the head surfaces. |
| Line 30: | Line 34: |
| {{attachment:meshMethods.JPG||height="400",width="350"}} | {{attachment:callAnat.gif}} |
| Line 32: | Line 36: |
| == iso2mesh == iso2mesh is a Matlab /octave-based mesh generation and processing toolbox. It can [[http://iso2mesh.sourceforge.net/cgi-bin/index.cgi?Doc/Workflow|create 3D tetrahedral finite element (FE) mesh from surfaces, 3D binary and gray-scale volumetric images]] such as segmented MRI/CT scans. |
== Iso2mesh == [[http://iso2mesh.sourceforge.net|Iso2mesh]] is a Matlab/Octave-based mesh generation and processing toolbox, available as a [[https://Tutorials/Plugins|Brainstorm plugin]]. |
| Line 35: | Line 39: |
| === Requirement === it If iso2mesh is not installed in your computer, Brainstrom will download the last release from this [[https://neuroimage.usc.edu/brainstorm/http://iso2mesh.sourceforge.net/cgi-bin/index.cgi?Download|webpage]] and install it when it is needed. However, you can also download the iso2mesh from the [[https://github.com/fangq/iso2mesh|github]] and add it to your Matlab path. |
Brainstorm uses it to generate a FEM tetrahedral mesh from a set of '''nested surfaces''' representing the separation between different tissues of the head. For example, these surfaces can be the ones generated for the computation of a [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#BEM_surfaces|BEM forward model]]. A full example is available in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/Duneuro#FEM_mesh|Realistic head model: FEM with DUNEuro]]. |
| Line 38: | Line 41: |
| === When and how to use it === iso2mesh is used as the basic option by brainstorm to generate FEM mesh from surfaces mesh. |
{{attachment:iso2meshOptions.gif}} |
| Line 41: | Line 43: |
| Assuming the situation where you have surfaces mesh of your subject available and you have already computed the [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem?highlight=(bem)|OpenMeeg]]forward problem. If you want to use the duneuro FEM to compute the forward model, you need to generate the FEM mesh from a similar surface used by OpenMeeg Here is the way to do it : | ==== Options ==== * '''MergeMesh''': Simply concatenates the input surfaces without any intersection checks. Default option (faster). * '''MergeSurf''': Concatenates and checks for intersections, split intersecting elements. Advanced option (slower). * '''Max tetrahedral volume''': Maximum volume of the tetrahedral element in the mesh. * From our tests, a DUNEuro FEM head model with a value of 0.1 achieves similar results as the OpenMeeg head model computed from the same surfaces. We have also noticed that the result with v = 0,001 is almost similar to v = 0,01. * Increasing the mesh resolution requires more time to generate the mesh, more time and memory to perform the FEM computation and more storage space in the database. * '''Percentage of elements kept''': Parameter between 0-100%, used to keep or not the original input surface nodes. |
| Line 43: | Line 51: |
| 1. Richt-click on the subject: In this way, brainstorm will load the inner, outer and the head from the subject data. if any of these surfaces is missing, an error will be displayed. 1. Select the 'Generate FEM mesh' item, 1. Select the iso2mesh option, 1. Set the iso2mesh parameters, |
==== Examples ==== * FEM mesh with different values of "Max volume": [10, 1, 0.1, 0.01] - Kept ratio=100%.<<BR>><<BR>> {{attachment:iso2meshMaxvol.gif}} * FEM meshes with only two compartments: This could be useful for investigating the influence of a specific tissue on the EEG/MEG forward solution or on the source localization, or for analyzing SEEG only within the brain volume. On the left: head and outer skull; On the right: inner and outer skull. <<BR>><<BR>> {{attachment:iso2Mesh2layer.gif}} |
| Line 48: | Line 55: |
| 1. These options are used by the surf2mesh function. Select either '''MergeMesh''' or '''MergeSurf.''' 1. '''Max tetrahedral volum''' : is the maximum volume of the tetrahedral element in the mesh. '''Pourcentage of the element to keep''': parameter between 0-100%, it used to keep or not the original input surface nodes. |
==== Troubleshooting ==== * '''Tetget failed''': If intersections are present on the surfaces mesh, the iso2mesh FEM mesh generation fails (tetgen). You may need regenerate new surfaces from the MRI. ''' ''' * Alternatively: You may try with the MergSurf option, this option can correct the intersection and create new nodes and elements. However, we do not recommend to use these models for EEG/MEG forward head computations: this is a research topic and it's still under investigation by the FEM communities. |
| Line 52: | Line 59: |
| Also, a full example is explained in this [[https://neuroimage.usc.edu/brainstorm/Duneuro?highlight=(duneuro)|page]]. | == Brain2mesh == [[http://mcx.space/brain2mesh/|Brain2mesh]] is a MATLAB/Octave based 3D mesh generation toolbox dedicated to the creation of high-quality multi-layered brain mesh models. This software is developed by the same team developing Iso2mesh and relies heavily on it. Both are available as a [[https://Tutorials/Plugins|Brainstorm plugins]]. Warning: Requires the '''Imaging Processing Toolbox'''. |
| Line 54: | Line 62: |
| . | Brainstorm runs the '''SPM12 '''segmentation routine on the '''T1 '''or '''T1+T2 MRI''' volumes to obtain a 5-tissue classification (white matter, gray matter, CSF, skull and skin), which is then passed to Brain2mesh for 3D meshing. A full example is available in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemTensors#FEM_mesh|FEM tensors estimation]]. This option runs fast, but does not generate good quality cortex surfaces, which are needed for the full cortically-constrained source estimation pipeline in Brainstorm. |
| Line 56: | Line 64: |
| {{attachment:iso2meshProcess.JPG||height="380",width="720"}} | At the moment, Brainstorm can only use the default parameters of Brain2mesh. If you need more options to be available from the interface, please contact us on the user forum. |
| Line 58: | Line 66: |
| Here is a view of mesh obtained with different values of the Max volume = [10, 1, 0.1, 0.01] with a keep ratio = 100%. | {{attachment:brain2meshCall.gif}} |
| Line 60: | Line 68: |
| {{attachment:IcbmMeshModels.jpg||height="420",width="700"}} | {{attachment:brain2meshMesh.gif}} |
| Line 62: | Line 70: |
| [[https://neuroimage.usc.edu/brainstorm/reviewMEEGForward?highlight=(duneuro)|From our tests]], a FEM head model with a value of 0,1 for the tetrahedral volume achieves similar results as the OpenMeeg head model computed from the same surfaces. We have also noticed that the result with v = 0,001 is almost similar to v = 0,01. | ==== Troubleshooting ==== * '''SPM-related errors''': If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: '''Restart Matlab''' to get a fresh workspace. |
| Line 64: | Line 73: |
| Increasing the mesh resolution needs more time to generate the mesh, more time to perform the FEM computation and of course more memory to store the mesh in the database. | == SimNIBS == [[https://simnibs.github.io/simnibs|SimNIBS]] software was developed to calculate the electric fields caused by Transcranial Electrical Stimulation (TES) and Transcranial Magnetic Stimulation (TMS). All its software dependencies (CAT12, Netgen, Gmsh, MeshFix) are embedded in the SimNIBS installation, which ensures a high stability and portability. However, it cannot be managed automatically as a plugin by Brainstorm, and needs to be installed manually. See: [[https://simnibs.github.io/simnibs/build/html/installation/simnibs_installer.html|Download and install SimNIBS]]. Two versions of SimNIBS are currently supported, that include different segmentation pipelines: SimNIBS 3.x (headreco) and SimNIBS 4.x (charm). |
| Line 66: | Line 76: |
| '''<<TAG(Advanced)>>''' | === SimNIBS 3: headreco === SimNIBS 3.x includes the pipeline [[https://simnibs.github.io/simnibs/build/html/documentation/command_line/headreco.html|headreco]] to process '''T1''' or '''T1+T2 MRI''' volumes (T1 is required, [[https://simnibs.github.io/simnibs/build/html/tutorial/head_meshing.html|T2 is recommended]]) and generate a high-quality tetrahedral FEM mesh. It calls internally [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]] for tissue segmentation (white matter, gray matter, CSF, skull and scalp), and therefore gives us access to high-quality cortex surfaces and surface-based atlases. The tetrahedral mesh generation is done with Gmsh, Netgen and MeshFix. Depending on your computer performances, this process will take between 2 to 5 hours. |
| Line 68: | Line 79: |
| If intersections are present on the surfaces mesh, the iso2mesh FEM mesh generation fails (tetgen) and an error will be displayed on the screen. If you face this problem, you need to check the surfaces and/or regenerate new surfaces from the MRI. | {{attachment:simnibs.gif}} |
| Line 70: | Line 81: |
| === other application === You can also select any surface mesh, or multiple surfaces (with Shift key), on the brainstorm anatomy windows and then generate tetrahedral mesh by following the same steps explained above. |
{{attachment:simnibs2.gif}} |
| Line 73: | Line 83: |
| Here are some examples using only 2 tissues. This option could be useful for investigation of tissues influence on the EEG/MEG on the forward solution or on the source localization, furthermore, this option could be used for analyzing only SEEG within brain volume. | {{attachment:simnibs3.png}} |
| Line 75: | Line 85: |
| {{attachment:otherMesh.JPG||height="320",width="600"}} | ==== Options ==== * '''Vertex density''': Number of node per mm2 of the surface mesh. * '''Number of vertices''': This is not an input parameter of SimNIBS, but a parameter to control how much to downsample the cortex surface generated by CAT12 when importing it into the Brainstorm database. See the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Import_the_anatomy|introduction tutorials]]. |
| Line 77: | Line 89: |
| == brain2mesh == Brain2Mesh is a MATLAB/Octave based 3D mesh generation toolbox dedicated to the creation of high-quality multi-layered brain mesh models. |
=== SimNIBS 4: charm === SimNIBS 4.x includes the pipeline [[https://simnibs.github.io/simnibs/build/html/tutorial/head_meshing.html#head-modeling-tutorial|charm]] to process '''T1''' or '''T1+T2 MRI''' volumes (T1 is required, [[https://simnibs.github.io/simnibs/build/html/tutorial/head_meshing.html|T2 is recommended]]). This pipeline generates the highest-quality tetrahedral FEM mesh we can get from Brainstorm. It creates segmentations and meshes with more tissues than SimNIBS3/headreco: white matter, gray matter, CSF, compact bone, spongy bone, scalp, eyes, blood and muscle. It also creates high-resolution cortex surfaces registered with the FSAverage template, but it does not generate the full CAT12 output folder anymore. For surface and volume-based atlases, it might be necessary to execute CAT12 from Brainstorm on top of SimNIBS. |
| Line 80: | Line 92: |
| === Requirement === Brain2Mesh is developed by the same team that developed the iso2mesh toolbox. Therefore iso2mesh is required. So if these toolboxes are not available on your computer, Brainstorm will download the last release and install it when it's needed. |
Given the high quality of its outputs, SimNIBS/charm is the recommended method for FEM mesh generation in Brainstorm. A full example is available in the tutorial: [[https://neuroimage.usc.edu/brainstorm/Tutorials/FemMedianNerveCharm#FEM_mesh_with_SimNIBS|FEM median nerve example]]. |
| Line 83: | Line 94: |
| You may also need the [[https://www.fil.ion.ucl.ac.uk/spm/software/spm12/|SPM12 toolbox]]. Brain2mesh is used only to generate tetrahedral mesh from the segmentation output. Therefore a segmentation of the MRI will be performed by SPM when this option is called. | {{attachment:charm1.gif}} |
| Line 85: | Line 96: |
| More parameters will be added in the next version. If you are using this method you can request our support to help you or to add these parameters asap. | {{attachment:charm2.png}} |
| Line 87: | Line 98: |
| === When and how to use it === This option is used when you have the individual MRI of the subject either T1 or T1 and T2. As said before, the SPM toolbox is required. The time required for this option is around 1 hour. here is the view of the obtained mesh from a T1 MRI |
==== Options ==== * '''Number of vertices''': This is not an input parameter of SimNIBS, but a parameter to control how much to downsample the cortex surface generated by CHARM when importing it into the Brainstorm database. See the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Import_the_anatomy|introduction tutorials]]. |
| Line 90: | Line 101: |
| {{attachment:brain2meshModel.JPG||height="450",width="600"}} | ==== Troubleshooting ==== * SimNIBS help: https://simnibs.github.io/simnibs/build/html/installation/throubleshooting.html * Forum: [[https://neuroimage.usc.edu/forums/t/brainstorm-not-recognising-path-or-command-to-simnibs-4-0-0/38963|SimNIBS library problems on Linux]] |
| Line 92: | Line 105: |
| This option is based on the [[http://mcx.space/brain2mesh/|brain2mesh toolbox]], we keep the default options. We will add more flexibility to control these options in the next future. However, if you want to use this option and you need support, we can help... just post your question on the forum or email us. | == ROAST == [[https://www.parralab.org/roast/|ROAST]] is a fully automated, Realistic, vOlumetric Approach to Simulate Transcranial electric stimulation. Open-source and Matlab-based, it is available as a [[https://Tutorials/Plugins|Brainstorm plugin]]. It calls internally SPM8 for tissue segmentation of the '''T1 '''or '''T1+T2 MRI''' volumes to obtain a 5-tissue classification (white matter, gray matter, CSF, skull and skin). Then it relies mostly on iso2mesh for generating a tetrahedral mesh. This option runs fast, but does not generate good quality cortex surfaces, which are needed for the full cortically-constrained source estimation pipeline in Brainstorm. {{attachment:roast1.gif}} {{attachment:roast2.gif}} ==== Troubleshooting ==== * '''SPM-related errors''': If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: '''Restart Matlab''' to get a fresh workspace. |
| Line 95: | Line 118: |
| This option call [[www.fieldtriptoolbox.org/tutorial/headmodel_eeg_fem/|the process of fieldtrip MRI segmentation]] (function ft_volumesegment) and hexahedral mesh generation (ft_meshprepare) develloped by the [[https://www.mrt.uni-jena.de/simbio/index.php?title=Main_Page|SimBio]] team. | [[http://www.fieldtriptoolbox.org/|FieldTrip]] is an open-source Matlab-based toolbox that includes a pipeline dedicated to the generation of [[http://www.fieldtriptoolbox.org/tutorial/headmodel_eeg_fem/|hexahedral FEM mesh]]. It is available as a [[https://Tutorials/Plugins|Brainstorm plugin]]. |
| Line 97: | Line 120: |
| === Requirement === To use this option, the [[http://www.fieldtriptoolbox.org/getting_started/|Fieldtrip]] and [[https://www.fil.ion.ucl.ac.uk/spm/software/spm12/|SPM toolbox]] should be in your matlab. |
Brainstorm calls the function [[https://github.com/fieldtrip/fieldtrip/blob/release/ft_volumesegment.m|ft_volumesegment]] on the '''T1 MRI''' volume to obtain a tissue segmentation with 5 layers (white matter, gray matter, CSF, skull and skin), then [[https://github.com/fieldtrip/fieldtrip/blob/release/ft_prepare_mesh.m|ft_prepare_mesh_hexa]] to create a hexahedral mesh. |
| Line 100: | Line 122: |
| === When and how to use it === This option can be called by two processes, either from the MRI or from any segmented tissue available on the brainstorm database. |
The mesh generation with the method is simple and fast: it downsamples the tissue classification volume, then converts all the voxels to a hexahedral mesh. The quality of the output is relatively poor: the regular meshing of the voxels makes it inefficient for providing a good representation of the tissues geometry with a limited number of elements. |
| Line 103: | Line 124: |
| The mesh generation with the method is faster. It converts all the voxels to hexahedral mesh. | {{attachment:fieldtrip1.gif}} |
| Line 105: | Line 126: |
| Only the hexahedral mesh is available for this method. You can either call this option from the MRI data or from any segmentation data available on the subject. If you call it from the MRI, a segmentation is processed first, then the mesh. If you call from the tissues, only the mesh process will be performed. | {{attachment:fieldtrip2.gif}} |
| Line 107: | Line 128: |
| {{attachment:mriAndTissue.JPG||height="450",width="700"}} | ==== Options ==== * '''Downsample volume before meshing''': When processing the tissue classification volume, reduce the number of voxels along each dimension by this factor. * '''Shift nodes to fit geometry''': The option calls the adaptative mesh generation. The process moves the nodes located on the interface either inward or outward in order to fit the geometry as explained in the [[http://www.fieldtriptoolbox.org/tutorial/headmodel_eeg_fem/|FieldTrip tutorial]]. Below, the example from the FieldTrip website, left=original, right=shifted.<<BR>><<BR>> {{attachment:nodeShiftFigure.JPG||height="200",width="500"}} |
| Line 109: | Line 132: |
| Right-click on the MRI (or the tissues), then "Generate FEM Mesh" then select Fieldtrip option. There are two parameters that the user needs to set, the downsampling of the volume and the node shift ratio. | ==== Troubleshooting ==== * '''SPM-related errors''': If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: '''Restart Matlab''' to get a fresh workspace. |
| Line 111: | Line 135: |
| {{attachment:fieldTripMeshCall.jpg||height="380",width="650"}} | == Zeffiro == [[https://github.com/sampsapursiainen/zeffiro_interface/wiki/Finite-Element-Mesh-generation|Zeffiro]] (ZI) is an open source code package constituting an accessible tool for multidisciplinary finite element (FE) based forward and inverse simulations in complex geometries, available as a [[https://Tutorials/Plugins|Brainstorm plugin]]. [TODO: Sampsa/Fernando: update the logo]. |
| Line 113: | Line 138: |
| The option "Downsamp volume before meshing" will reduce the number of voxel by this factor. | Please note that Zeffiro requires the Parallel Computing Toolbox. For further information, please see [[https://github.com/sampsapursiainen/zeffiro_interface/wiki/Finite-Element-Mesh-generation|Zeffiro]] pages. |
| Line 115: | Line 140: |
| The "Shift node" option calls the adaptative mesh generation. The process moves the nodes located on the interface either inward or outward in order to fit the geometry as explained [[http://www.fieldtriptoolbox.org/tutorial/headmodel_eeg_fem/|here]]. This figure shows an example (from Fieldtrip webpage), left the unshifted and on the right the shifted. | {{attachment:ZefMeshFig0.jpg||height="306",width="324"}} |
| Line 117: | Line 142: |
| {{attachment:nodeShiftFigure.JPG||height="200",width="500"}} | Brainstorm uses the Zeffiro Mesh tool to generate a FEM tetrahedral mesh from a set of '''nested surfaces''' representing the separation between different tissues of the head. For example, these surfaces can be generated to compute a [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutBem#BEM_surfaces|BEM forward model]]. |
| Line 119: | Line 144: |
| This method is fast compare to the previous options, the following figures show examples of the mesh obtained with fieldtrip option from the ICBM model. | Zeffiro Mesh module plugin in Brainstorm follows the same structure as the iso2mesh case. Users can call the mesh generation either from the Subject node (fig1) or a set of selected surfaces (fig2). To select multiple surfaces, you can hold the "Ctrl" key and then select multiple surfaces with the mouse. |
| Line 121: | Line 146: |
| {{attachment:fieldTripMeshICBM.JPG||height="300",width="700"}} | If the user selects the subject node, Brainstorm automatically detects the three main surfaces [head, outer, and inner skull]. [TODO: we can extend this for more surfaces if required] |
| Line 123: | Line 148: |
| == SimNIBS == [[https://simnibs.github.io/simnibs/build/html/index.html|SimNIBS]]software develloped to calculate electric fields caused by Transcranial Electrical Stimulation (TES) and Transcranial Magnetic Stimulation (TMS). From its pipline, brainstorm integrates the process of the automatic segmentation of MRI images and meshing to create individualized head models. This process is called "headreco and it's explained [[https://simnibs.github.io/simnibs/build/html/documentation/command_line/headreco.html|here]]. |
{{attachment:ZefMeshFig1.jpg||height="750",width="750"}} |
| Line 126: | Line 150: |
| === Requirement === SimNibs is an independat software, brainstorm call its functions internally thereore you need to install SimNibs and its depemdencies as explained in [[https://simnibs.github.io/simnibs/build/html/installation/simnibs_installer.html|this webpage]]. |
If the user selects multiple surfaces [hold the Ctrl keyboard and select the surfaces with the mouse], Brainstorm will use all the selected surfaces to generate the FEM mesh. |
| Line 129: | Line 152: |
| This method used the SimNibs software. So to call this process, you need to download and install the SimNibs software, the process of the installation is explained in the SimNibs webpage : https://simnibs.github.io/simnibs/build/html/installation/simnibs_installer.html. | {{attachment:ZefMeshFig2.jpg||height="630",width="630"}} |
| Line 131: | Line 154: |
| When you have installed SimNibs, Brainstorm can call the main function used for the mesh generation frm the main graphical interface. Depemding on your computer performances, this process will take between 2 to 5 hours. We highly recommend to close all other running process and application on our computer in order to speed this process. | Once the Zeffiro option is selected, Brainstorm asks the user to either use the advanced Zeffiro mesh module [click on "Yes"] or use the basic options from Brainstorm (Click on "NO" Recommended). |
| Line 133: | Line 156: |
| 1- Create new subject within the current protocole | Please refer to the Zeffiro documentation [TODO: Sampsa/Fernando] for the advanced options. |
| Line 135: | Line 158: |
| 2- Load the T1 of the subject to the brainstorm database. | For the basic options, Brainstorm will ask the users to set two options: the mesh resolution as the length edge of the mesh elements in (mm); the second option asking for using the GPU computation, as shown in the following figure. |
| Line 137: | Line 160: |
| 3- Associate a T2 mri to the subject if it's available (this is better for csf/skull/scalp segmentation) | {{attachment:ZefMeshFig3.jpg||height="420",width="420"}} |
| Line 139: | Line 162: |
| 4- Right-click on the subject, select the "Generate FEM mesh" | The following figure illustrates the FEM mesh obtained using the Zef Mesh module on a defaced head surface, outer skull, and inner skull. |
| Line 141: | Line 164: |
| . Select "SIMNIBS", and choose the "Tetrahedral element" and keep the other options to the default value. | {{attachment:ZefMeshFig4.jpg||height="420",width="420"}} |
| Line 143: | Line 166: |
| The headreco function is fully integrated to brainstorm. With this option, brainstorm can reconstruct a tetrahedral head mesh from T1- and T2-weighted structural MR images. It runs also with only a T1w image, but it will achieve more reliable skull segmentations when a T2w image is supplied. | == On the hard drive == Right-click on a FEM mesh > File > View file contents: |
| Line 145: | Line 169: |
| https://www.fil.ion.ucl.ac.uk/spm/software/spm12/ | {{attachment:femContents.gif}} |
| Line 147: | Line 171: |
| == Tissue anisotropy estimation == === Brainsuite Installation [TODO] === === Volume generation from surface files === '''<<TAG(Advanced)>>''' === Volume generation from T1/T2 MRI data === You can also generate your own FEM head model and then load it to brainstorm. However the automatic head model generation from from imaging techniques are not accurate and most of the time visual checking are needed and manual correction are required. ==> this depends lagely on the quality of the T1/T2 MRI image(https://simnibs.github.io/simnibs/build/html/tutorial/head_meshing.html). This step is based on the "roast" toolbox (link to roast : https://github.com/andypotatohy/roast ) that we adapted for the MEEG forward computation. If you want to generate your own FEM head model from an MRI, you will need to download these file (link), then run the bst process as explained here. * If there is a MRI file with the string "T2" in the subject anatomy folder, it will use it * Otherwise, if you select explicitly two MRI files with CTRL+Click, it will use the first one as the T1 and the second one as the T2 (this needs to be documented in the tutorial) === FEM Head model template === - Load the FEM volumic mesh (template created from ICBM T1 MRI using SimNibs) - Load the surface mesh (template created also from ICBM using ICBM ) and then generates the volume mesh (either tetra or hexa) by calling the tetgen process cia iso2mesh toolbox (if hexa are desired, the tetra mesh will be converted to hexa ... ) https://github.com/brainstorm-tools/brainstorm3/issues/185#issuecomment-576749612 === Head model based on the level set approach === TODO and Validate |
==== Structure of the FEM mesh files: tess_fem_*.mat ==== * '''Comment''': String displayed in the database explorer to represent the file. * '''Vertices''': [Nvertices x 3], coordinates (x,y,z) of all the tetrahedral or hexahedral mesh nodes in SCS coordinates. * '''Elements''': * Tetrahedral mesh: [Nelements x 4], list of indices corresponding to the rows of the Vertices matrix, each set of 4 connected vertices identifying a tetrahedron of the mesh. * Hexahedral mesh: [Nelements x 8], list of indices corresponding to the rows of the Vertices matrix, each set of 8 connected vertices identifying a hexahedron of the mesh. * '''Tissue''': [Nelements x 1], for each Element, index of the corresponding tissue (e.g. wm=1, gm=2, csf=3, skull=4, scalp=5) * '''TissueLabels''': Labels of the tissues, e.g. {'white' 'gray' 'csf' 'skull' 'scalp'} * '''Tensors''': [Nelements x 12] if tensors are computed, otherwise empty. The 12 values are the eigenvalues and eigenvectors interpolated on each element of the mesh. * '''History''': List of operations performed on this file (menu File > View file history). |
FEM mesh generation
Authors: Takfarinas Medani, Francois Tadel
FEM forward modeling requires the construction of a 3D model of thee head tissues. The volume of the head is divided in small geometrical elements with 4 faces (tetrahedrons) or 6 faces (hexahedrons). Each element is associated with a type of biological tissue (e.g. white matter, gray matter, CSF, skull, skin) and electrical conductivity properties.
This page lists the methods integrated with Brainstorm to generate 3D meshes of the head. For a generic introduction to FEM in Brainstorm, refer to the tutorials: Realistic head model: FEM with DUNEuro and FEM median nerve example.
Contents
Generate FEM mesh
FEM meshes can be computed from surfaces (as the ones generated for the BEM models) or from MRI volumes (T1w and/or T2w). The methods that are available when using the popup menu Generate FEM mesh depend on the selected inputs.
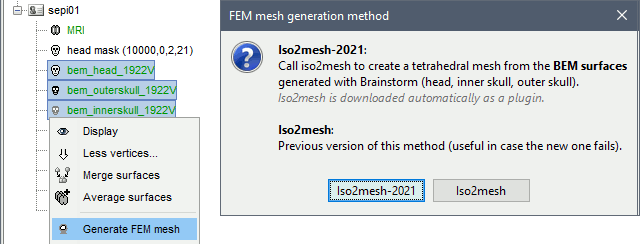
Surfaces
Select a list of surfaces representing the separation between different tissues (holding the CTRL or SHIFT key), then right-click on any of them. The software Iso2mesh can create a tetrahedral mesh to represent the tissues between these different layers.
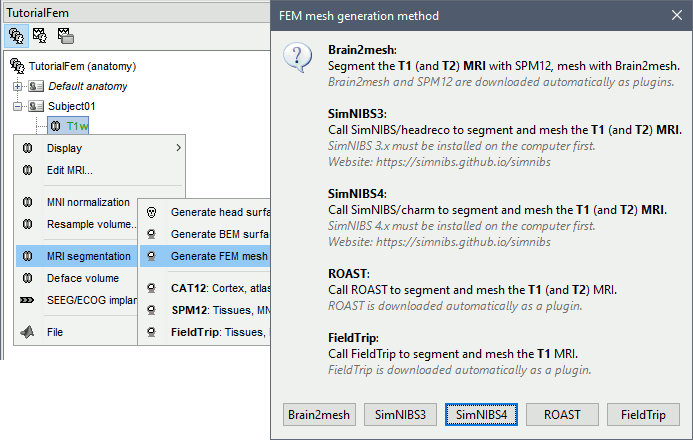
T1 MRI
Right-click on a T1 MRI available in the database. Typically, this is the default MRI volume displayed in green in the subject folder. Methods available: Brain2mesh, SimNIBS3, SimNIBS4, ROAST, FieldTrip.
T1+T2 MRI
Select the T1+T2 volumes, then right-click on any of them. The different files are identified based on the tags "T1" and "T2" the file names (as displayed in the Brainstorm database explorer). If these identification tags are not found in the file names, the default MRI (in green) is used as the T1, the other as the T2. If none of the is the default MRI, the first selected file is used as the T1, the second is used as the T2. Methods available: Brain2mesh, SimNIBS3, SimNIBS4, ROAST.
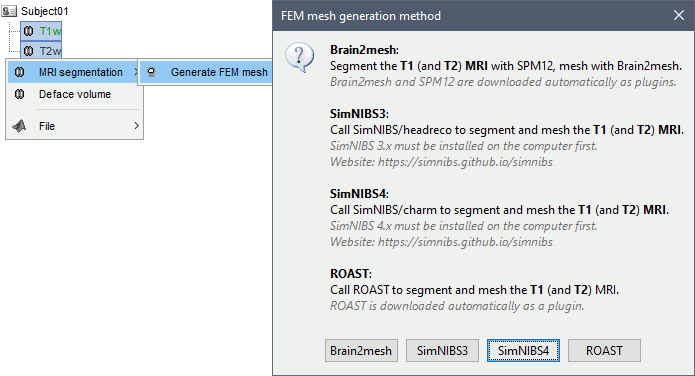
Anatomy folder
If you right-click on the subject folder > Generate FEM mesh, then Brainstorm offers all the possible options, even the ones that are not applicable to this specific subject.
Volume method: If using a method based on MRI volumes, the T1 and T2 volumes are detected among all the volumes available based on the tags "T1" and "T2" in the file names (make sure only one file as each of these tags), otherwise use only the default MRI (in green) as the T1.
Surface method: If using a method based on surfaces, the default surfaces (in green) from three categories as selected: the inner skull, the outer skull and the head surfaces.
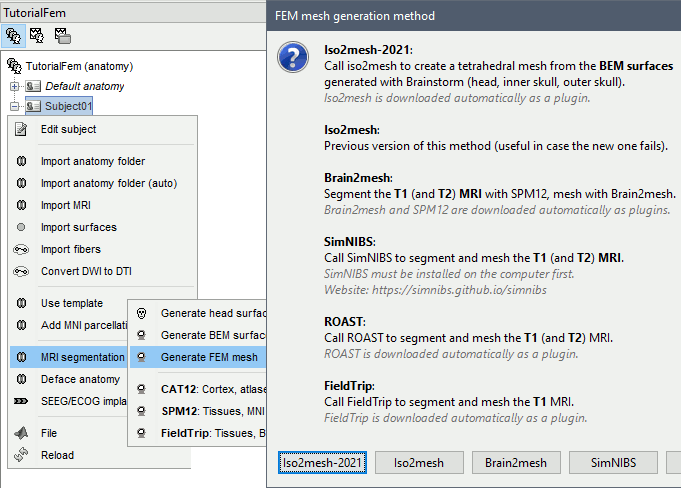
Iso2mesh
Iso2mesh is a Matlab/Octave-based mesh generation and processing toolbox, available as a Brainstorm plugin.
Brainstorm uses it to generate a FEM tetrahedral mesh from a set of nested surfaces representing the separation between different tissues of the head. For example, these surfaces can be the ones generated for the computation of a BEM forward model. A full example is available in the tutorial Realistic head model: FEM with DUNEuro.
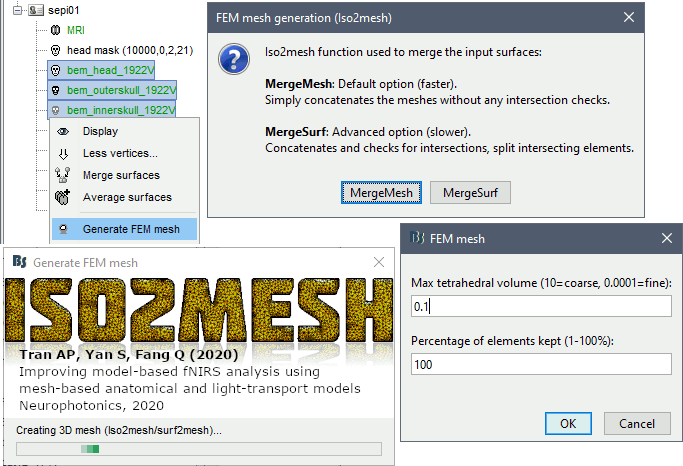
Options
MergeMesh: Simply concatenates the input surfaces without any intersection checks. Default option (faster).
MergeSurf: Concatenates and checks for intersections, split intersecting elements. Advanced option (slower).
Max tetrahedral volume: Maximum volume of the tetrahedral element in the mesh.
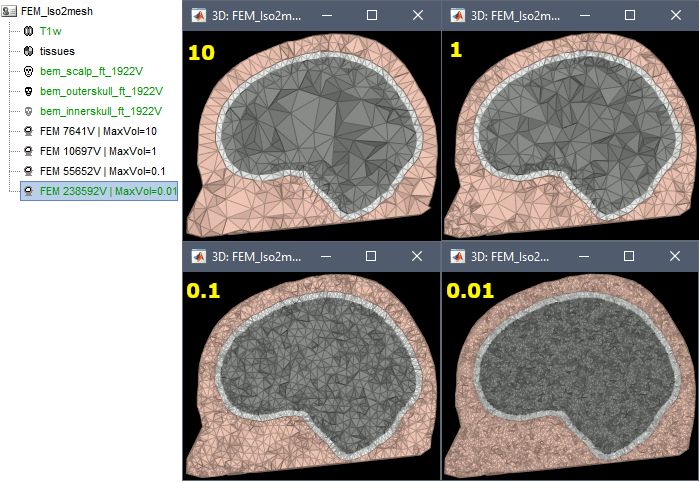
From our tests, a DUNEuro FEM head model with a value of 0.1 achieves similar results as the OpenMeeg head model computed from the same surfaces. We have also noticed that the result with v = 0,001 is almost similar to v = 0,01.
- Increasing the mesh resolution requires more time to generate the mesh, more time and memory to perform the FEM computation and more storage space in the database.
Percentage of elements kept: Parameter between 0-100%, used to keep or not the original input surface nodes.
Examples
FEM mesh with different values of "Max volume": [10, 1, 0.1, 0.01] - Kept ratio=100%.
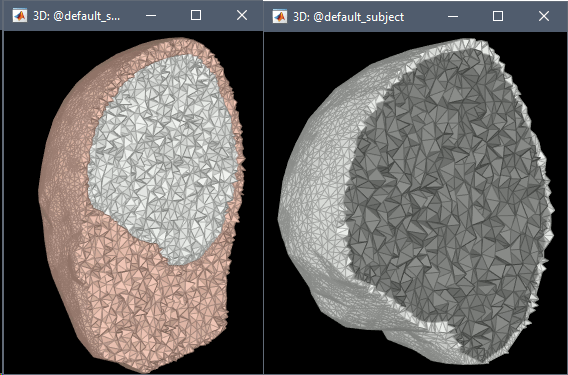
FEM meshes with only two compartments: This could be useful for investigating the influence of a specific tissue on the EEG/MEG forward solution or on the source localization, or for analyzing SEEG only within the brain volume. On the left: head and outer skull; On the right: inner and outer skull.
Troubleshooting
Tetget failed: If intersections are present on the surfaces mesh, the iso2mesh FEM mesh generation fails (tetgen). You may need regenerate new surfaces from the MRI.
Alternatively: You may try with the MergSurf option, this option can correct the intersection and create new nodes and elements. However, we do not recommend to use these models for EEG/MEG forward head computations: this is a research topic and it's still under investigation by the FEM communities.
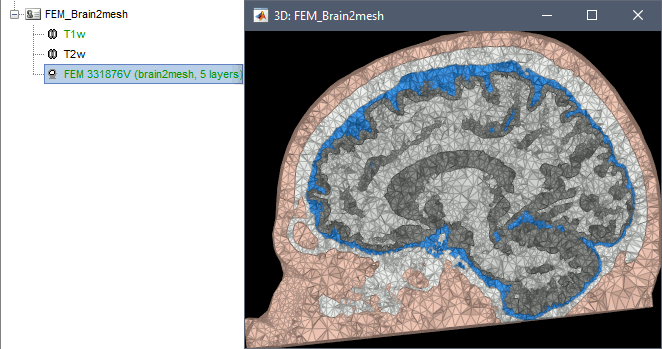
Brain2mesh
Brain2mesh is a MATLAB/Octave based 3D mesh generation toolbox dedicated to the creation of high-quality multi-layered brain mesh models. This software is developed by the same team developing Iso2mesh and relies heavily on it. Both are available as a Brainstorm plugins. Warning: Requires the Imaging Processing Toolbox.
Brainstorm runs the SPM12 segmentation routine on the T1 or T1+T2 MRI volumes to obtain a 5-tissue classification (white matter, gray matter, CSF, skull and skin), which is then passed to Brain2mesh for 3D meshing. A full example is available in the tutorial FEM tensors estimation. This option runs fast, but does not generate good quality cortex surfaces, which are needed for the full cortically-constrained source estimation pipeline in Brainstorm.
At the moment, Brainstorm can only use the default parameters of Brain2mesh. If you need more options to be available from the interface, please contact us on the user forum.
Troubleshooting
SPM-related errors: If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: Restart Matlab to get a fresh workspace.
SimNIBS
SimNIBS software was developed to calculate the electric fields caused by Transcranial Electrical Stimulation (TES) and Transcranial Magnetic Stimulation (TMS). All its software dependencies (CAT12, Netgen, Gmsh, MeshFix) are embedded in the SimNIBS installation, which ensures a high stability and portability. However, it cannot be managed automatically as a plugin by Brainstorm, and needs to be installed manually. See: Download and install SimNIBS. Two versions of SimNIBS are currently supported, that include different segmentation pipelines: SimNIBS 3.x (headreco) and SimNIBS 4.x (charm).
SimNIBS 3: headreco
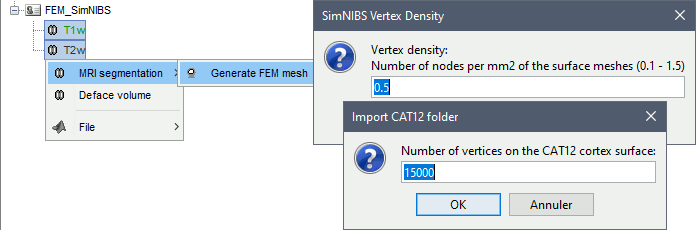
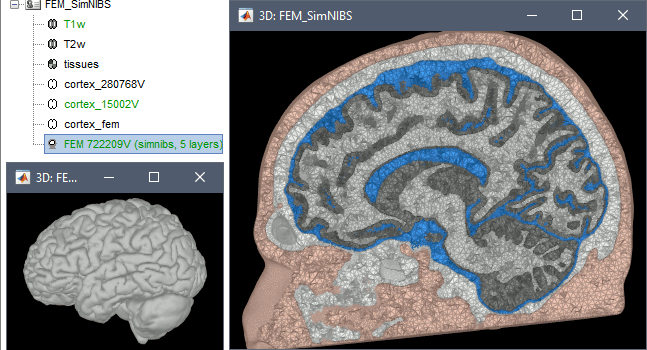
SimNIBS 3.x includes the pipeline headreco to process T1 or T1+T2 MRI volumes (T1 is required, T2 is recommended) and generate a high-quality tetrahedral FEM mesh. It calls internally CAT12 for tissue segmentation (white matter, gray matter, CSF, skull and scalp), and therefore gives us access to high-quality cortex surfaces and surface-based atlases. The tetrahedral mesh generation is done with Gmsh, Netgen and MeshFix. Depending on your computer performances, this process will take between 2 to 5 hours.
Options
Vertex density: Number of node per mm2 of the surface mesh.
Number of vertices: This is not an input parameter of SimNIBS, but a parameter to control how much to downsample the cortex surface generated by CAT12 when importing it into the Brainstorm database. See the introduction tutorials.
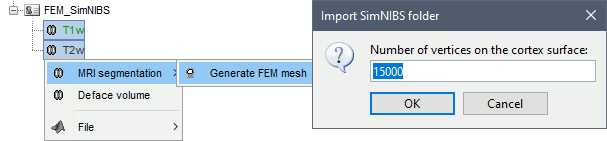
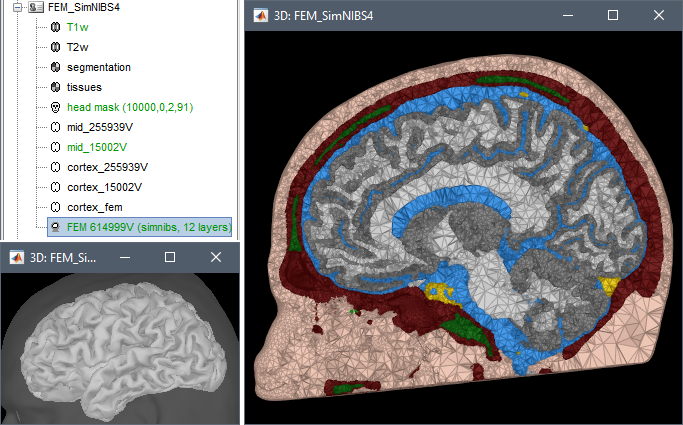
SimNIBS 4: charm
SimNIBS 4.x includes the pipeline charm to process T1 or T1+T2 MRI volumes (T1 is required, T2 is recommended). This pipeline generates the highest-quality tetrahedral FEM mesh we can get from Brainstorm. It creates segmentations and meshes with more tissues than SimNIBS3/headreco: white matter, gray matter, CSF, compact bone, spongy bone, scalp, eyes, blood and muscle. It also creates high-resolution cortex surfaces registered with the FSAverage template, but it does not generate the full CAT12 output folder anymore. For surface and volume-based atlases, it might be necessary to execute CAT12 from Brainstorm on top of SimNIBS.
Given the high quality of its outputs, SimNIBS/charm is the recommended method for FEM mesh generation in Brainstorm. A full example is available in the tutorial: FEM median nerve example.
Options
Number of vertices: This is not an input parameter of SimNIBS, but a parameter to control how much to downsample the cortex surface generated by CHARM when importing it into the Brainstorm database. See the introduction tutorials.
Troubleshooting
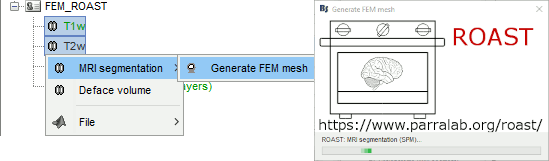
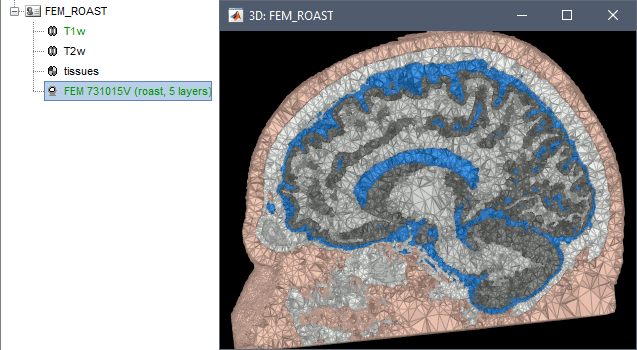
ROAST
ROAST is a fully automated, Realistic, vOlumetric Approach to Simulate Transcranial electric stimulation. Open-source and Matlab-based, it is available as a Brainstorm plugin. It calls internally SPM8 for tissue segmentation of the T1 or T1+T2 MRI volumes to obtain a 5-tissue classification (white matter, gray matter, CSF, skull and skin). Then it relies mostly on iso2mesh for generating a tetrahedral mesh.
This option runs fast, but does not generate good quality cortex surfaces, which are needed for the full cortically-constrained source estimation pipeline in Brainstorm.
Troubleshooting
SPM-related errors: If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: Restart Matlab to get a fresh workspace.
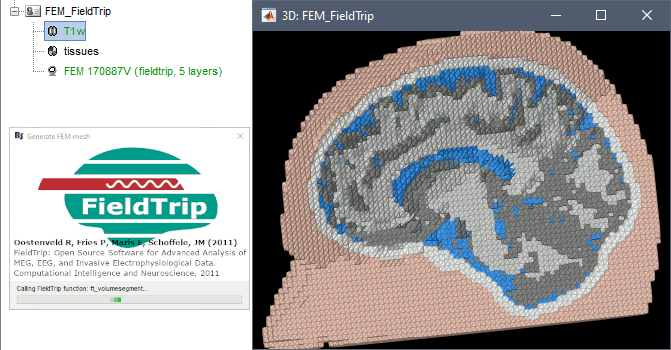
Fieldtrip
FieldTrip is an open-source Matlab-based toolbox that includes a pipeline dedicated to the generation of hexahedral FEM mesh. It is available as a Brainstorm plugin.
Brainstorm calls the function ft_volumesegment on the T1 MRI volume to obtain a tissue segmentation with 5 layers (white matter, gray matter, CSF, skull and skin), then ft_prepare_mesh_hexa to create a hexahedral mesh.
The mesh generation with the method is simple and fast: it downsamples the tissue classification volume, then converts all the voxels to a hexahedral mesh. The quality of the output is relatively poor: the regular meshing of the voxels makes it inefficient for providing a good representation of the tissues geometry with a limited number of elements.
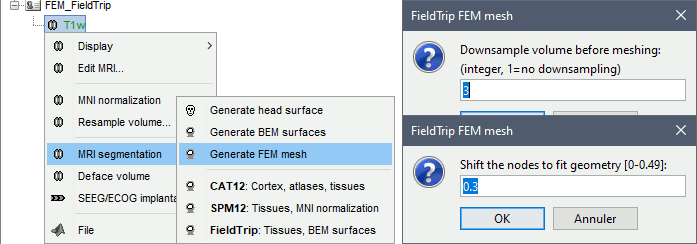
Options
Downsample volume before meshing: When processing the tissue classification volume, reduce the number of voxels along each dimension by this factor.
Shift nodes to fit geometry: The option calls the adaptative mesh generation. The process moves the nodes located on the interface either inward or outward in order to fit the geometry as explained in the FieldTrip tutorial. Below, the example from the FieldTrip website, left=original, right=shifted.
Troubleshooting
SPM-related errors: If you've been trying multiple methods successively, errors mentioning a spm_*.m function could be due to incompatible versions of SPM12 functions in the Matlab path. Brain2mesh, FieldTrip and ROAST all run different versions of SPM12 from the same instance of Matlab. Solution: Restart Matlab to get a fresh workspace.
Zeffiro
Zeffiro (ZI) is an open source code package constituting an accessible tool for multidisciplinary finite element (FE) based forward and inverse simulations in complex geometries, available as a Brainstorm plugin. [TODO: Sampsa/Fernando: update the logo].
Please note that Zeffiro requires the Parallel Computing Toolbox. For further information, please see Zeffiro pages.

Brainstorm uses the Zeffiro Mesh tool to generate a FEM tetrahedral mesh from a set of nested surfaces representing the separation between different tissues of the head. For example, these surfaces can be generated to compute a BEM forward model.
Zeffiro Mesh module plugin in Brainstorm follows the same structure as the iso2mesh case. Users can call the mesh generation either from the Subject node (fig1) or a set of selected surfaces (fig2). To select multiple surfaces, you can hold the "Ctrl" key and then select multiple surfaces with the mouse.
If the user selects the subject node, Brainstorm automatically detects the three main surfaces [head, outer, and inner skull]. [TODO: we can extend this for more surfaces if required]
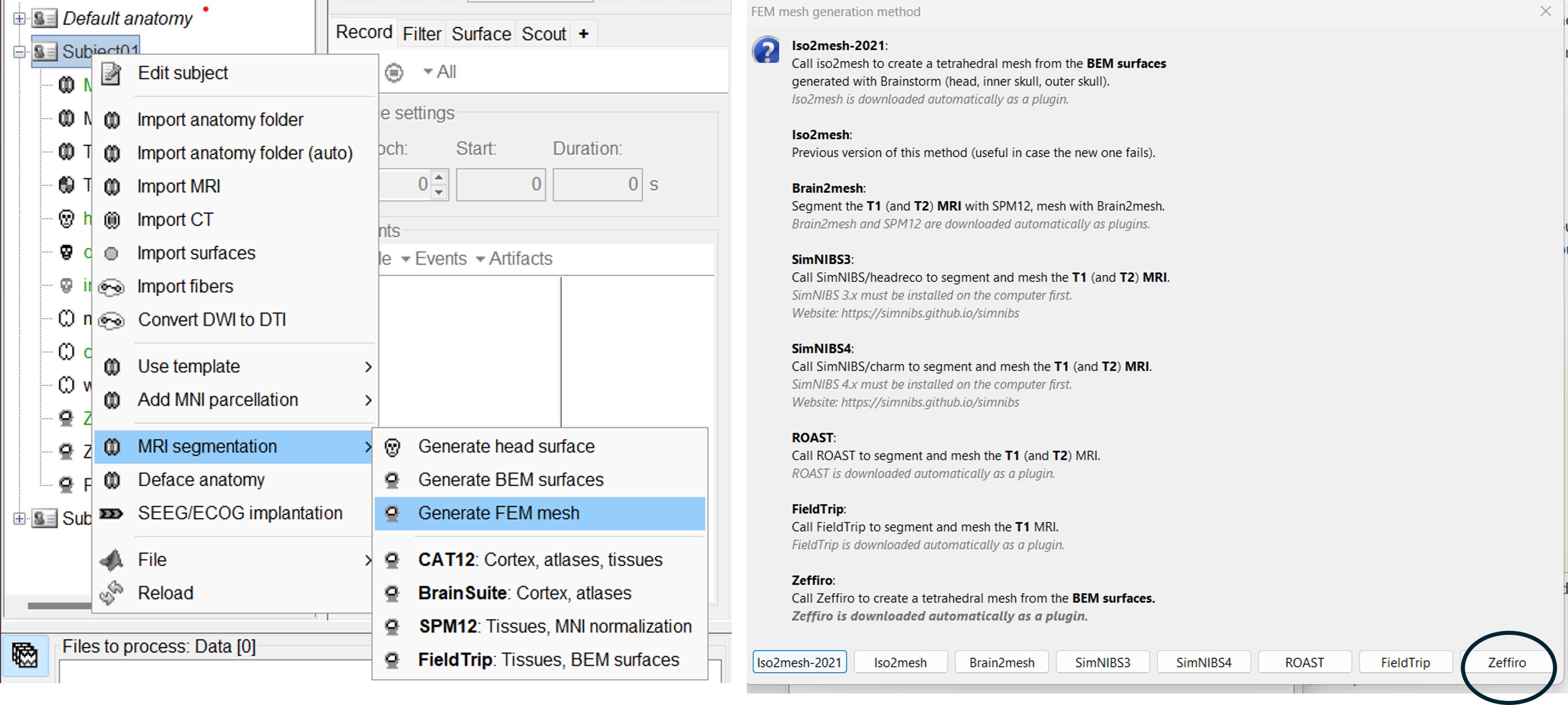
If the user selects multiple surfaces [hold the Ctrl keyboard and select the surfaces with the mouse], Brainstorm will use all the selected surfaces to generate the FEM mesh.
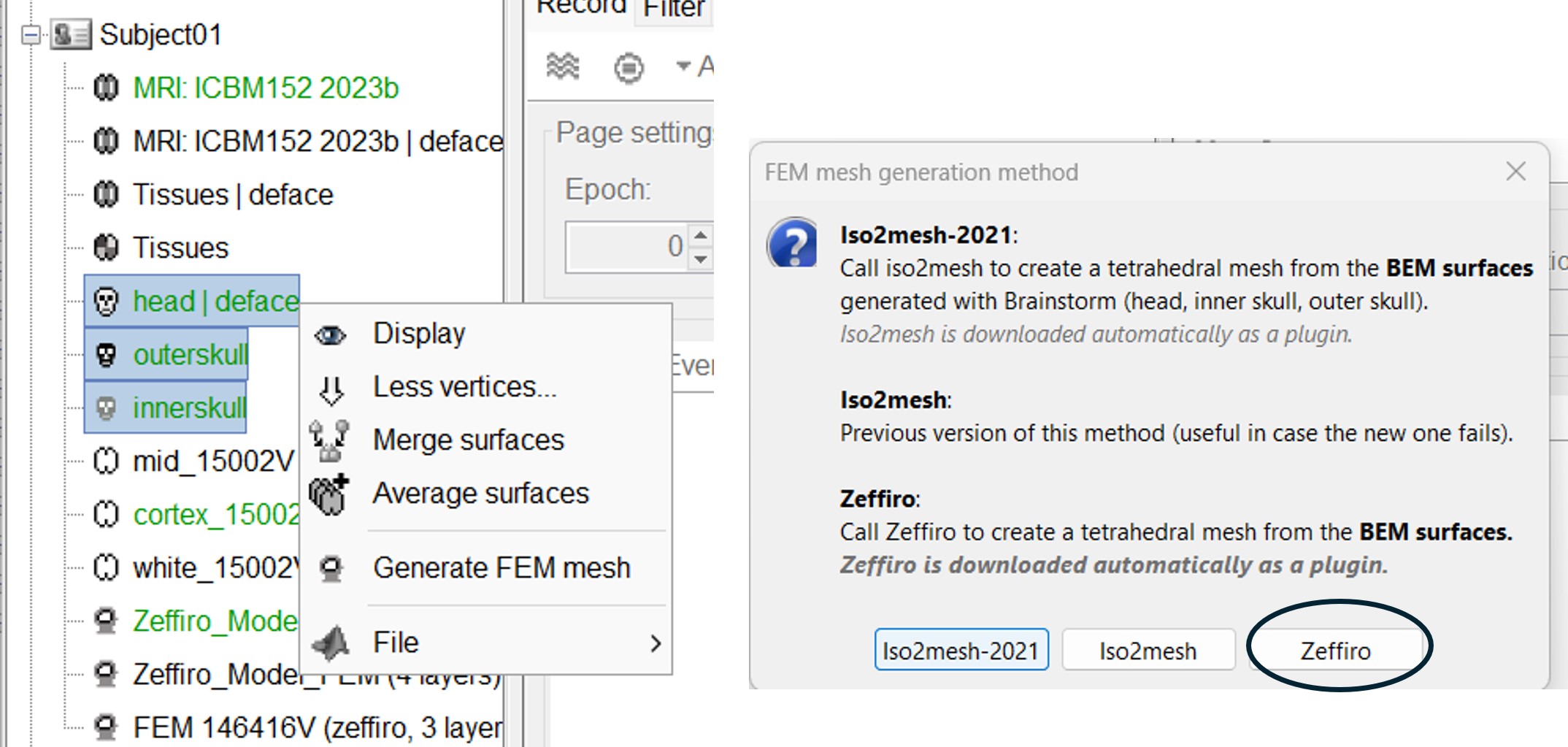
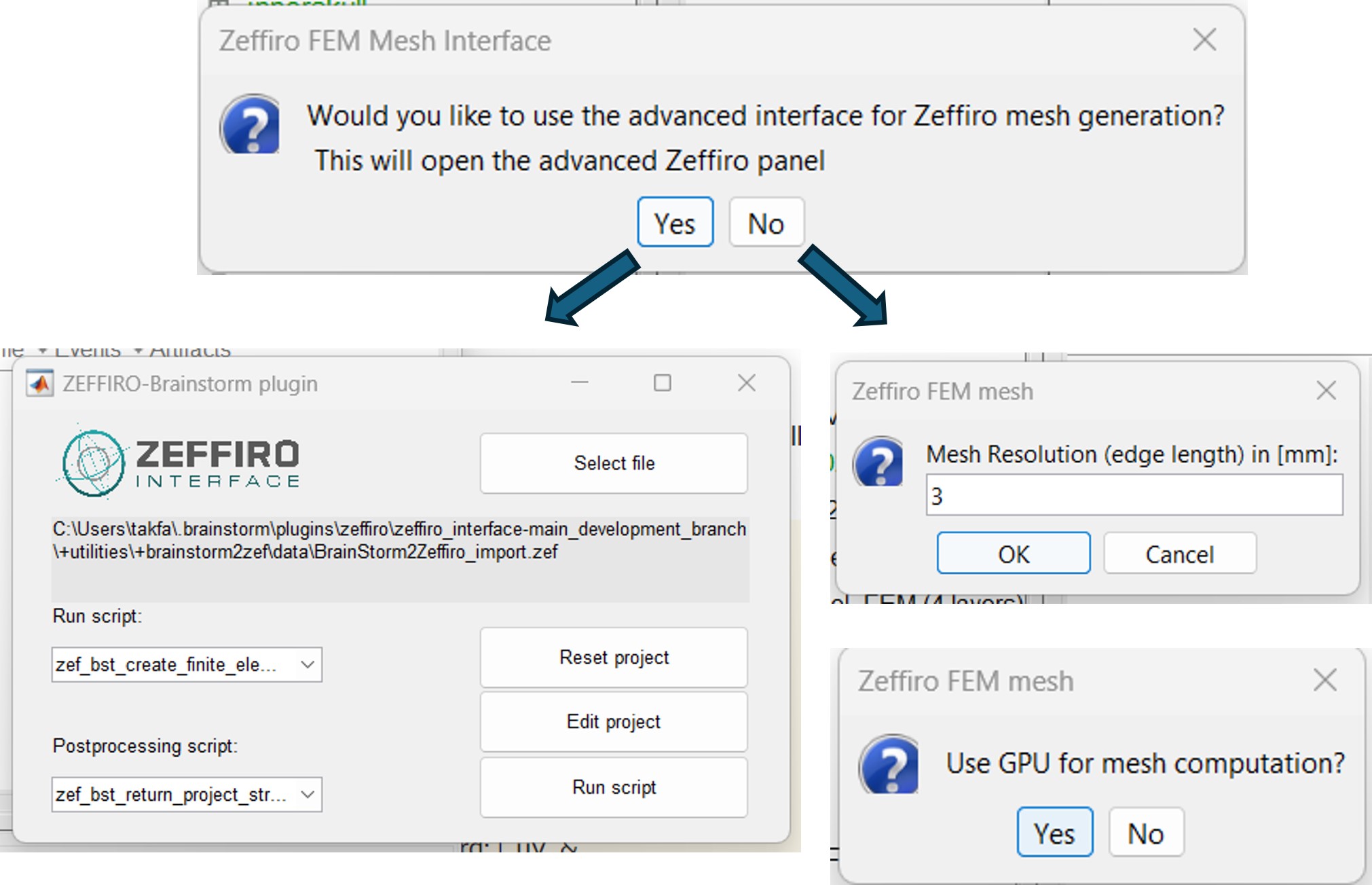
Once the Zeffiro option is selected, Brainstorm asks the user to either use the advanced Zeffiro mesh module [click on "Yes"] or use the basic options from Brainstorm (Click on "NO" Recommended).
Please refer to the Zeffiro documentation [TODO: Sampsa/Fernando] for the advanced options.
For the basic options, Brainstorm will ask the users to set two options: the mesh resolution as the length edge of the mesh elements in (mm); the second option asking for using the GPU computation, as shown in the following figure.
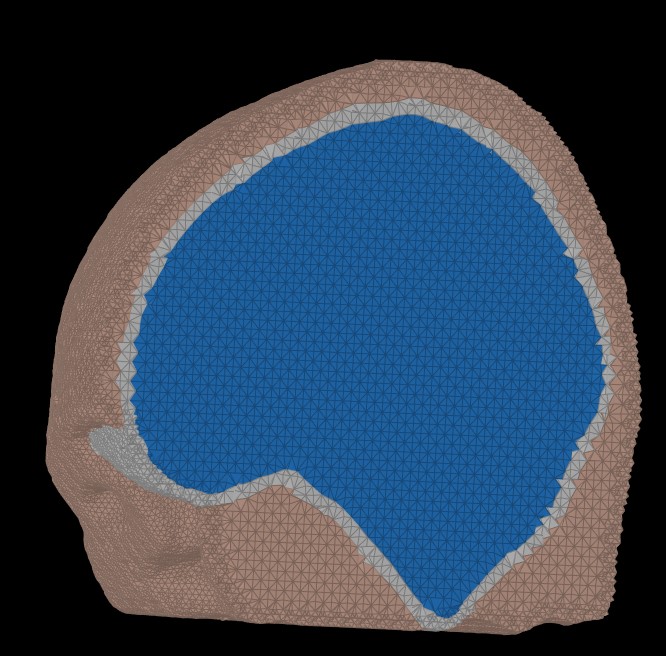
The following figure illustrates the FEM mesh obtained using the Zef Mesh module on a defaced head surface, outer skull, and inner skull.
On the hard drive
Right-click on a FEM mesh > File > View file contents:
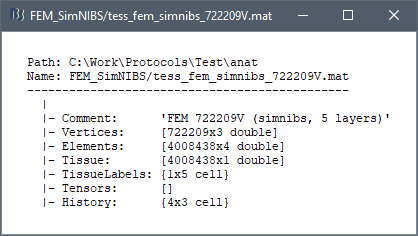
Structure of the FEM mesh files: tess_fem_*.mat
Comment: String displayed in the database explorer to represent the file.
Vertices: [Nvertices x 3], coordinates (x,y,z) of all the tetrahedral or hexahedral mesh nodes in SCS coordinates.
Elements:
- Tetrahedral mesh: [Nelements x 4], list of indices corresponding to the rows of the Vertices matrix, each set of 4 connected vertices identifying a tetrahedron of the mesh.
- Hexahedral mesh: [Nelements x 8], list of indices corresponding to the rows of the Vertices matrix, each set of 8 connected vertices identifying a hexahedron of the mesh.
Tissue: [Nelements x 1], for each Element, index of the corresponding tissue (e.g. wm=1, gm=2, csf=3, skull=4, scalp=5)
TissueLabels: Labels of the tissues, e.g. {'white' 'gray' 'csf' 'skull' 'scalp'}
Tensors: [Nelements x 12] if tensors are computed, otherwise empty. The 12 values are the eigenvalues and eigenvectors interpolated on each element of the mesh.
History: List of operations performed on this file (menu File > View file history).
