|
Size: 18321
Comment:
|
Size: 19873
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 20: Head model = | = Tutorial 20: Head modelling = |
| Line 4: | Line 4: |
| The following tutorials describe how brain activity can be estimated from the MEG/EEG recordings we have processed so far. This step consists in solving two separate modeling problems: the modeling of the electromagnetic properties of the head and of the sensor array (a.k.a. '''head model''' or '''forward model''') and the estimation of the brain sources which have produced the data. That second step is known as '''source modeling''' or solving an '''inverse problem'''. It requires that the forward modeling of the head tissues and sensor instrumentation is completed first. This tutorial explains how to compute a head model for the subject of the auditory oddball experiment. As far as source modeling is concerned, we focus the tutorial on linear estimates of distributed source models, which are popular and physiologically plausible approaches (there is no dipole fitting available in the software): you may want to refer to [[http://www.canada-meg-consortium.org/EN/MegIntro|other sources]] for a complete description of source imaging with MEG and EEG. '' '' | The following tutorials describe how brain activity can be estimated from the MEG/EEG recordings we have processed so far. This step consists in solving two separate modeling problems: the modeling of the electromagnetic properties of the head and of the sensor array (a.k.a. '''head model''' or '''forward model''') and the estimation of the brain sources which have produced the data. That second step is known as '''source modeling''' or solving an '''inverse problem'''. It requires that the forward modeling of the head tissues and sensor instrumentation is completed first. This tutorial explains how to compute a head model for the subject of the auditory oddball experiment.'' '' |
| Line 13: | Line 13: |
| If one of your primary objectives is to '''identify and map''' the regions of the brain involved in a specific stimulus response or behaviour, source estimation can help address this aspect. Empirical interpretations of sensor topographies can inform where about brain generators might be located: which hemisphere, what broad aspect of the anatomy (e.g., frontal vs. posterior regions). Source estimation methods improves anatomical resolution with respect to the interpreration of sensor patterns. Spatial resolution in MEG and EEG depends on source depth and orientation and overall SNR: still, one can expect to be able to source activations within the millimeter to centimeter range, especially relatively, when contrasts between conditions are implemented in the study design. '' '' | If one of your primary objectives is to '''identify and map''' the regions of the brain involved in a specific stimulus response or behaviour, source estimation can help address this aspect. Empirical interpretations of sensor topographies can inform where about brain generators might be located: which hemisphere, what broad aspect of the anatomy (e.g., frontal vs. posterior regions). Source estimation methods improves anatomical resolution with respect to the interpreration of sensor patterns. Spatial resolution in MEG and EEG depends on source depth and orientation and overall SNR: still, one can expect to be able to observe source activations within the millimeter to centimeter range, especially relatively, when contrasts between conditions are implemented in the study design. '' '' |
| Line 15: | Line 15: |
| Source mapping is a form of '''spatial''' '''deconvolution''' of sensor data. In EEG in particular, scalp topographies are very smooth and it is common to that contributions from distant brain regions overalp over large clusters of electrodes. Moving to the source space can help separating the active regions. '' '' | Source mapping is a form of '''spatial''' '''deconvolution''' of sensor data. In EEG in particular, scalp topographies are very smooth and it is common that contributions from distant brain regions overalp over large clusters of electrodes. Moving to the source space can help separating the active regions. '' '' |
| Line 17: | Line 17: |
| More specifically in '''MEG''', source maps can be a great assest to alleviate some specific issues with that modality. In MEG, the head of the subject is not fixed. Hence sensor topographies depend on the actual position of the subject under the rigid helmet. Therefore, between two acquisition runs, or between subjects with different head shapes and sizes, '''the same''' '''MEG sensors may pick up signals from different parts of the brain'''. This problem does not exist in EEG, where electrode montages are attached to the head and arranged according to standard positions. '' '' | More specifically in '''MEG''', source maps can be a great asset to alleviate some specific issues with that modality. In MEG, the head of the subject is not fixed. Hence sensor topographies depend on the actual position of the subject under the rigid helmet. Therefore, between two acquisition runs, or between subjects with different head shapes and sizes, '''the same''' '''MEG sensors may pick up signals from different parts of the brain'''. This problem does not exist in EEG, where electrode montages are attached to the head and arranged according to standard positions. '' '' |
| Line 19: | Line 19: |
| Another important point to consider when interpreting MEG sensor maps is that every MEG manufacturer uses different types of sensor technology (e.g., magnetometers vs. gradiometers; axial vs. tangential gradiometers, etc. yielding different physical measures). This is not an issue with EEG, with essentially one sensor type (electrodes, dry or active, all measuring Volts): two EEG electrode caps with same number of electrodes only differ in how and well they ensure good contact with the scalp. Working in source space alleviates all these aspects. '' '' | Another important point to consider when interpreting MEG sensor maps is that MEG manufacturers use different types of sensor technology (e.g., magnetometers vs. gradiometers; axial vs. tangential gradiometers, etc. yielding different physical measures). This is not an issue with EEG, with essentially one sensor type (electrodes, dry or active, all measuring Volts): two EEG electrode caps with same number of electrodes only differ in how well they ensure good contact with the scalp. Working in source space alleviates all these aspects. '' '' |
| Line 21: | Line 21: |
| Nevertheless, if your neuroscience question can be solved by measuring signal latencies, or other aspects which do not depend crucially on anatomical localization (such as in global signal properties integrated over all or clusters of sensors), source modeling might not be a requirement. To sort out this question wil influence the time and hardware requirements to complete your data analysis (source analysis multiplies the needs in terms of disk storage and computational requirements). | Nevertheless, if your neuroscience question can be solved by measuring signal latencies over broad regions, or other aspects which do not depend crucially on anatomical localization (such as global signal properties integrated over all or clusters of sensors), source modeling might not be a requirement. To sort out this question will influence the time and hardware requirements to complete your data analysis (source analysis multiplies the needs in terms of disk storage and computational specifications). |
| Line 25: | Line 25: |
| == The origins of the MEG/EEG signals == To understand how we are reconstructing the sources, it is interesting to have an idea of the physiological origins of MEG and EEG signals. The models we use are based on some physiological assumptions that are not always valid, understanding them may help you in selecting the appropriate method. '' '' |
== The origins of MEG/EEG signals == To better understand how the forward model is elaborated, we need to have at least a basic understanding of the physiological origins of MEG/EEG signals. Note that, as always when dealing with modeling, we need to deal with various degrees of approximation. '' '' |
| Line 28: | Line 28: |
| It is assumed that most of the currents we record are related with the postsynaptic activity of the pyramidal neurons in the cerebral cortex. These cells are '''aligned spatially''' and '''perpendicular to the cortex surface'''. Millions of post-synaptic potentials in the apical dendrites of neighbouring pyramidal neurons sum up in time and space to form what we can approximate at a macroscopic level with a few electric dipoles (green arrows below). '' '' | Overall, it is assumed that most of the MEG/EEG signals are generated by postsynaptic activity of ensembles of cortical pyramidal neurons of the cerebral cortex. The reason is essentially in the morphology and mass effect of these cells, which present '''elongated shapes '''and are '''oriented''' '''perpendicularly to the cortical surface'''. Mass effects of close-to-simulatenous changes in post-synaptic potentials pyramidal neural assemblies add up in time and space. These effects can conveniently be modeled at a mesoscopic spatial scale with electric dipoles distributed along the cortical mantle (green arrows in figure below). Note that there is growing evidence that MEG and EEG are also sensitive to deeper, cortical and subcortical structures, including brain nuclei and the cerebellum, where pyramidal cells are rare or absent. Brainstorm features advanced models that can include as an option, these structures. The emphasis in the present tutorial is on pyramidal cell assemblies for simplicity. |
| Line 30: | Line 30: |
| The primary and volume currents generated by these dipoles create a potential distribution and a magnetic field at the surface of the head. We can record them with bipolar montages of electrodes placed on the skin (EEG) or very sensitive superconducting detectors (SQUIDs/MEG). '' '' | The primary and volume currents generated by these dipoles create differences in electrical potentials and magnetic fields that can be detected outside the head. They can be measured with electrodes placed on the skin (EEG, with respect to a reference) or very sensitive magnetic detectors (MEG). '' '' |
| Line 38: | Line 38: |
| '''Dipole fitting vs distributed methods''' '' '' | '''Dipole fitting vs distributed models''' |
| Line 40: | Line 40: |
| The source estimation process consists in estimating the position and activity of a set of electric dipoles, to approximate the activity of the brain that produced the MEG/EEG data we recorded. Two families of solutions were explored in the past decades: the '''dipole fitting methods''' (we estimate the position and amplitude of a very limited number of dipoles over short time windows) and the '''distributed methods''' (we define a priori a dense grid of dipoles and then estimate their activity from the recordings). '' '' | MEG/EEG source estimation consists in modeling brain activity with current dipoles. A current dipole is a convenient model equivalent to the net electrophysiological activity of local assemblies of neurons. Two main approaches have been explored for source MEG/EEG estimation: '''dipole fitting methods''' - where the position and amplitude of a one to a few equivalent current dipoles (ECD) are estimated over relativelyshort time windows - and '''distributed models''' - where the location (and typically, the orientation) of a large number dipoles is fixed. The dipoles sample a spatial grid covering the entire brain volume or the cortical surface - only their amplitude is estimated at each time point. '' '' |
| Line 42: | Line 42: |
| The single dipole fitting approaches are very efficient in specific cases where we know in advance the number or regions involved and their latencies of activity. But they are difficult to generalize and automate, and not adapted for group analysis. With Brainstorm, we decided to work only with distributed source models, which require less manual tuning for getting acceptable results. '' '' | Equivalent dipole fitting approaches are quite straightforward and can be adequate when the number of brain regions expected to be active is small. Therefore, it is most adequate for responses at early latencies. They cannot generalize to capture complex dynamics over extended period of time (epochs) and the associated estimation techniques are quite sensitive to intial conditions (how many dipoles to fit? Where does the serch start? etc.). Our strategy in Brainstorm is to promote distributed source models, which are less user-sensitive, can generalize to all experimental conditions, and yield time-resolved image volumes that can be processed in many different, powerful ways (group statistics, spatial segmentation, use of regions fo interest, correspondence with fMRI, etc.) |
| Line 46: | Line 46: |
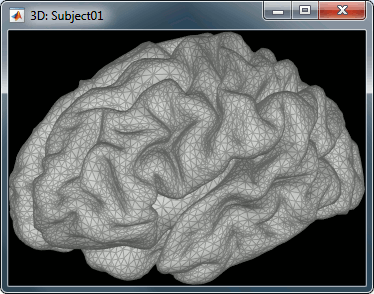
| Our first step of modeling consists in defining the '''positions and orientations of the dipoles''' for which we want to estimate the activity. This set of dipoles is our '''source space'''. By default, we limit our analysis to the '''cortex envelope''', based on this observation that most of the MEG/EEG signals is related with the synchronous activity of assemblies or cortical pyramidal cells. The simple solution we recommend is to directly use the vertices of the cortical surface we imported in the first tutorials (the nodes we can see in the grey mesh in the left image below). '' '' | When opting for distributed source models, the '''positions and orientations of the elementary dipoles''' that will define the "voxel" grid of the source images produced need to be defined. This set of dipoles is called the '''source space'''. By default, Brainstorm constrains the source space to the '''cortex''', where signal-to-noise and sensitivity is maximum in MEG/EEG. Note however that more complete models that include subcortical structures and the cerebellum are available in Brainstorm. Therefore, one decision you need to make before proceeding with source imaging is whether more complete source spaces are required to answer your neuroscience question. |
| Line 48: | Line 48: |
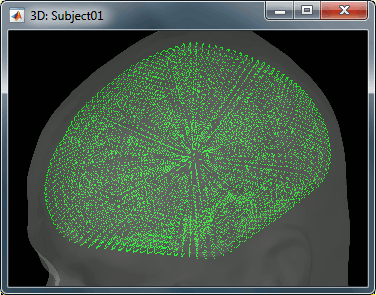
| In order to represent all the possible dipole orientations, we define '''three dipoles for each vertex''' of the cortex surface, corresponding to three orthogonal directions (X,Y,Z). When importing the anatomy of the subject, we downsampled the cortex surface to '''15,000 vertices'''. This will correspond to a source space of '''45,000 dipoles'''. We will compute a forward model that connects the activity of these 45,000 dipoles with the 275 MEG sensors we have in this dataset. '' '' | For this tutorial, we use the simple approach where a current dipole is automatically assigned to each of the vertices of the cortical surface we have imported so far (see the nodes in the grey mesh in the leftmost image below). '' ''Also in this turorial, we elected not to contrain the dipole orientations to be perpendicular to the cortical surface. This choice can be motivated by the fact that you have no access to the individual MRI of your participant and thefore resort to a generic template, or that you find the cortical orientation constraint to be poorly unjustified. In this context, '''three orthogonal dipoles '''are assigned to each vertex of the cortex. This triplet forms a basis to account for local currents flowing in any arbitrary direction, not just orthogonal to the cortex. When importing the anatomy of the subject, we downsampled the cortex surface to '''15,000 vertices'''. This will correspond to a source space of '''45,000 dipoles'''. We will compute a forward model that connects the activity of these 45,000 dipoles with the 275 MEG sensors we have in this dataset. '' '' |
| Line 138: | Line 142: |
| * External documentation: [[http://www.canada-meg-consortium.org/EN/MegBaillet5|Electromagnetic neural source imaging]] |
Tutorial 20: Head modelling
Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Richard Leahy, Sylvain Baillet
The following tutorials describe how brain activity can be estimated from the MEG/EEG recordings we have processed so far. This step consists in solving two separate modeling problems: the modeling of the electromagnetic properties of the head and of the sensor array (a.k.a. head model or forward model) and the estimation of the brain sources which have produced the data. That second step is known as source modeling or solving an inverse problem. It requires that the forward modeling of the head tissues and sensor instrumentation is completed first. This tutorial explains how to compute a head model for the subject of the auditory oddball experiment.
Contents
Why estimating sources?
Reconstructing the activity of the brain from MEG or EEG recordings involves several sophisticated steps. Athough Brainstorm simplifies the procedures required, it is important to understand whether source modeling is essential to answer the neuroscience question which brought you to collect data in the first place.
If one of your primary objectives is to identify and map the regions of the brain involved in a specific stimulus response or behaviour, source estimation can help address this aspect. Empirical interpretations of sensor topographies can inform where about brain generators might be located: which hemisphere, what broad aspect of the anatomy (e.g., frontal vs. posterior regions). Source estimation methods improves anatomical resolution with respect to the interpreration of sensor patterns. Spatial resolution in MEG and EEG depends on source depth and orientation and overall SNR: still, one can expect to be able to observe source activations within the millimeter to centimeter range, especially relatively, when contrasts between conditions are implemented in the study design.
Source mapping is a form of spatial deconvolution of sensor data. In EEG in particular, scalp topographies are very smooth and it is common that contributions from distant brain regions overalp over large clusters of electrodes. Moving to the source space can help separating the active regions.
More specifically in MEG, source maps can be a great asset to alleviate some specific issues with that modality. In MEG, the head of the subject is not fixed. Hence sensor topographies depend on the actual position of the subject under the rigid helmet. Therefore, between two acquisition runs, or between subjects with different head shapes and sizes, the same MEG sensors may pick up signals from different parts of the brain. This problem does not exist in EEG, where electrode montages are attached to the head and arranged according to standard positions.
Another important point to consider when interpreting MEG sensor maps is that MEG manufacturers use different types of sensor technology (e.g., magnetometers vs. gradiometers; axial vs. tangential gradiometers, etc. yielding different physical measures). This is not an issue with EEG, with essentially one sensor type (electrodes, dry or active, all measuring Volts): two EEG electrode caps with same number of electrodes only differ in how well they ensure good contact with the scalp. Working in source space alleviates all these aspects.
Nevertheless, if your neuroscience question can be solved by measuring signal latencies over broad regions, or other aspects which do not depend crucially on anatomical localization (such as global signal properties integrated over all or clusters of sensors), source modeling might not be a requirement. To sort out this question will influence the time and hardware requirements to complete your data analysis (source analysis multiplies the needs in terms of disk storage and computational specifications).
The origins of MEG/EEG signals
To better understand how the forward model is elaborated, we need to have at least a basic understanding of the physiological origins of MEG/EEG signals. Note that, as always when dealing with modeling, we need to deal with various degrees of approximation.
Overall, it is assumed that most of the MEG/EEG signals are generated by postsynaptic activity of ensembles of cortical pyramidal neurons of the cerebral cortex. The reason is essentially in the morphology and mass effect of these cells, which present elongated shapes and are oriented perpendicularly to the cortical surface. Mass effects of close-to-simulatenous changes in post-synaptic potentials pyramidal neural assemblies add up in time and space. These effects can conveniently be modeled at a mesoscopic spatial scale with electric dipoles distributed along the cortical mantle (green arrows in figure below). Note that there is growing evidence that MEG and EEG are also sensitive to deeper, cortical and subcortical structures, including brain nuclei and the cerebellum, where pyramidal cells are rare or absent. Brainstorm features advanced models that can include as an option, these structures. The emphasis in the present tutorial is on pyramidal cell assemblies for simplicity.
The primary and volume currents generated by these dipoles create differences in electrical potentials and magnetic fields that can be detected outside the head. They can be measured with electrodes placed on the skin (EEG, with respect to a reference) or very sensitive magnetic detectors (MEG).
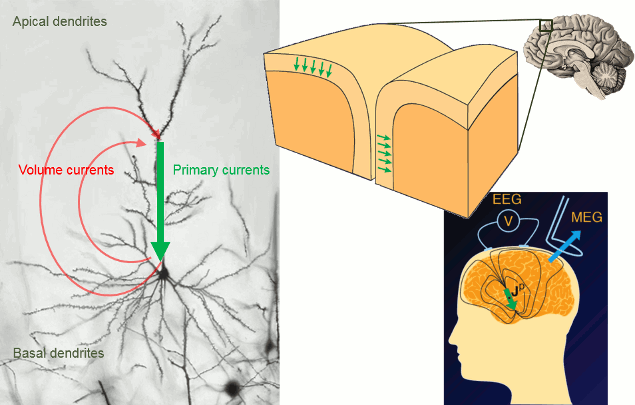
- Matti Hamalainen, 2007
Source space
Dipole fitting vs distributed models
MEG/EEG source estimation consists in modeling brain activity with current dipoles. A current dipole is a convenient model equivalent to the net electrophysiological activity of local assemblies of neurons. Two main approaches have been explored for source MEG/EEG estimation: dipole fitting methods - where the position and amplitude of a one to a few equivalent current dipoles (ECD) are estimated over relativelyshort time windows - and distributed models - where the location (and typically, the orientation) of a large number dipoles is fixed. The dipoles sample a spatial grid covering the entire brain volume or the cortical surface - only their amplitude is estimated at each time point.
Equivalent dipole fitting approaches are quite straightforward and can be adequate when the number of brain regions expected to be active is small. Therefore, it is most adequate for responses at early latencies. They cannot generalize to capture complex dynamics over extended period of time (epochs) and the associated estimation techniques are quite sensitive to intial conditions (how many dipoles to fit? Where does the serch start? etc.). Our strategy in Brainstorm is to promote distributed source models, which are less user-sensitive, can generalize to all experimental conditions, and yield time-resolved image volumes that can be processed in many different, powerful ways (group statistics, spatial segmentation, use of regions fo interest, correspondence with fMRI, etc.)
Location constraints
When opting for distributed source models, the positions and orientations of the elementary dipoles that will define the "voxel" grid of the source images produced need to be defined. This set of dipoles is called the source space. By default, Brainstorm constrains the source space to the cortex, where signal-to-noise and sensitivity is maximum in MEG/EEG. Note however that more complete models that include subcortical structures and the cerebellum are available in Brainstorm. Therefore, one decision you need to make before proceeding with source imaging is whether more complete source spaces are required to answer your neuroscience question.
For this tutorial, we use the simple approach where a current dipole is automatically assigned to each of the vertices of the cortical surface we have imported so far (see the nodes in the grey mesh in the leftmost image below). Also in this turorial, we elected not to contrain the dipole orientations to be perpendicular to the cortical surface. This choice can be motivated by the fact that you have no access to the individual MRI of your participant and thefore resort to a generic template, or that you find the cortical orientation constraint to be poorly unjustified.
In this context, three orthogonal dipoles are assigned to each vertex of the cortex. This triplet forms a basis to account for local currents flowing in any arbitrary direction, not just orthogonal to the cortex.
When importing the anatomy of the subject, we downsampled the cortex surface to 15,000 vertices. This will correspond to a source space of 45,000 dipoles. We will compute a forward model that connects the activity of these 45,000 dipoles with the 275 MEG sensors we have in this dataset.
This default number of 15,000 vertices is empirical. Over the years, our experience seemed to show that it represents a good balance between the representation of the brain circumvolutions, the surface sampling and the amount of data that is generated. Using less vertices makes it difficult to preserve the shape of the brain, using more vertices produces more data without adding to the spatial resolution of the method and may lead to computational memory issues.
Orientation constraints
Additionally, we can impose constraints of orientation on the dipoles, to match the physiological observation that the pyramidal cells are mostly organized perpendicularly to the cortex surface. This has the advantage of limiting the number of dipoles to 15,000 (one per vertex) and making the results much easier to display and process. However, this constraint is most of the time too strong and distorts the reconstruction. This orientation constraint is an option of the inverse model and will be discussed in the following introduction tutorials.
Fully unconstrained
The spatial constraint of imposing all the dipoles to be on the cortical surface might also be too restrictive in some cases, because our model is then not able to correctly represent the activity in deeper brain structures or in the cerebellum. Therefore we also offer an option to use the entire brain volume as our source space (the green dots below represent dipoles locations in volume model). This produces results that can be better or worse depending on the data, but in all the cases much more difficult to review. Volume and mixed head volumes are discussed in the advanced tutorials about source modeling.
Forward problem
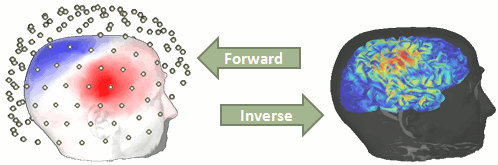
The first step of the source reconstruction consists in computing a model that explains how the electric currents or the magnetic fields flow from the electric generators in the brain (source space) through the different tissues of the head (mostly brain, CSF, skull and skin), to finally reach the sensors.
The question of building a model that connects the values we observe outside of the head (MEG/EEG) to the electric activity of the cortical dipoles in the brain is called forward problem.
The model we obtain after solving this problem is called head model in Brainstorm, but can also be referred to as forward model, leadfield matrix or gain matrix.
In this tutorial we will use the default source space: the low-resolution cortex surface with 15,000 vertices, as the support of 45,000 dipoles. We will use indifferently the terms dipole and source.
What we expect to get at the end of this process is a matrix [Nsensors x Nsources].
Available methods for MEG forward modeling
Single sphere: The head is considered as a homogeneous sphere.
Overlapping spheres: Refines the previous model by fitting one local sphere for each sensor.
OpenMEEG BEM: Symmetric Boundary Element Method from the open-source software OpenMEEG. Described in an advanced tutorial: BEM head model.
Models recommended for each modality
MEG: Overlapping spheres.
The magnetic fields are not affected too much by the heterogeneity of the tissues of the head. There is no real need for modeling the head with too much detail.EEG: OpenMEEG BEM.
The electric currents are strongly affected by jumps between very conductive tissues (brain, CSF, skin) and an isolant medium (the skull). A realistic head model is advised for integrating the properties of the skull correctly. When computing a BEM model is not an option, for instance if OpenMEEG crashes for unknown reasons, the Berg's three-layer sphere can be an acceptable option.sEEG/ECoG: The OpenMEEG BEM option is the only one available.
Computation
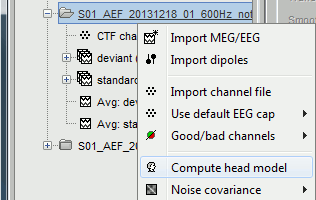
The forward models are related with the anatomy of the subject and the description of the sensors, therefore the menus associated to its computation are attached to the channel file.
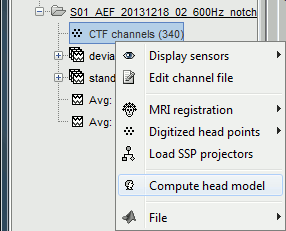
In the imported Run#01, right-click on the channel file or the folder > Compute head model.
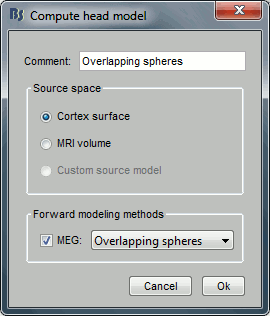
Keep the default options selected: Source space=Cortex, Forward model=Overlapping spheres.
You obtain one new file in the database. It is always saved in the same folder as the channel file.
There is not much you can do with this file except for using it for estimating sources. This will be the purpose of the following tutorials.Right-click on the new head model > Check spheres. This window shows the spheres that were estimated. You can check them by following the indications written in green at the bottom of the window: use left/right arrows. At each step, the current sensor marker is displayed in red, and the sphere you see is its local estimation of the inner skull shape.
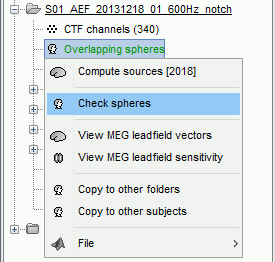
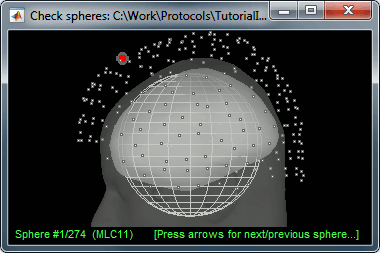
This algorithm is supposed to use the inner skull surface from the subject, but we usually do not have this information. In this case, a pseudo-innerskull is reconstructed using a dilated version of the cortex envelope.
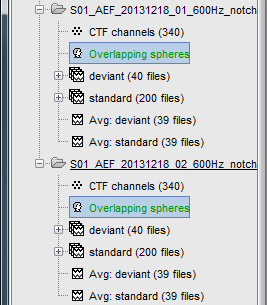
Repeat the same operation for the other file. We have two different acquisition runs with two different relative position of the head and the sensors, therefore we need to compute two different head models.
In the imported Run#02, right-click on the channel file > Compute head model.
Database explorer
Additional considerations about the management of the head model files.
If you have multiple head models computed in the same folder, you would see one displayed in green and the others in black. The one in green is selected as the default head model, it will be used for all the following computation steps. To change the default selection, double-click on another head model file (or right-click > Set as default head model).
You can use the database explore for batching the computation of the head model. The menu "Compute head model" is available in popup menus in the database explorer at all the levels. It is applied recursively to all the folders contained in the node(s) you selected.
On the hard drive
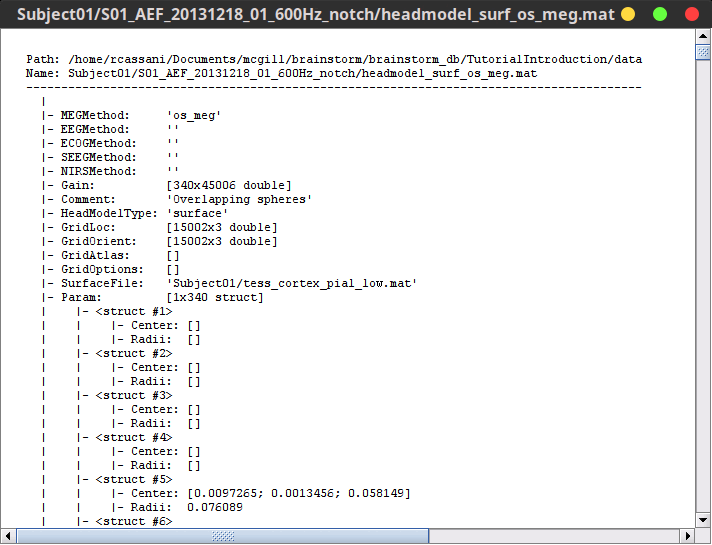
Right-click on any head model > File > View file contents:
Structure of the head model files
MEGMethod: Forward model used for MEG sensors ('os_meg', 'meg_sphere', 'openmeeg' or empty).
EEGMethod: Forward model used for EEG sensors ('eeg_3sphereberg', 'openmeeg' or empty).
ECOGMethod: Forward model used for ECoG sensors ('openmeeg' or empty).
SEEGMethod: Forward model used for sEEG sensors ('openmeeg' or empty).
Gain: Leadfield matrix, [Nsensors x Nsources], equivalent to [Nsensors x 3*Nvertices]
Comment: String displayed in the database explorer to represent this file.
HeadModelType: Type of source space used for this head model ('surface', 'volume', 'mixed').
GridLoc: [Nvertices x 3], (x,y,z) positions of the grid of source points. In the case of a surface head model, it corresponds to a copy of the 'Vertices' matrix from the cortex surface file.
GridOrient: [Nvertices x 3], direction of the normal to the surface for each vertex point (copy of the 'VertNormals' matrix of the cortex surface). Empty in the case of a volume head model.
GridAtlas: In the case of mixed head models, contains a copy of the "Source model options" atlas structure that was used for creating the model.
SurfaceFile: Relative path to the cortex surface file related with this head model.
Param: Description of the sphere that was estimated for each sensor (Center/Radius).
Gain matrix
The Gain matrix stores the leadfield for 3 orientations (x,y,z) at each grid point (p1, p2, ...).
The successive columns of the Gain matrix are: [p1_x, p1_y, p1_z, p2_x, p2_y, p2_z ...]To convert this unconstrained leadfield matrix to a constrained model, where the orientation of each dipole is fixed and normal to the cortex surface:
Export the head model file to the HeadModel structure: Right-click > File > Export to Matlab.
> Gain_constrained = bst_gain_orient(HeadModel.Gain, HeadModel.GridOrient);
The dimension of the output matrix is three times smaller: [Nsensors x Nvertices]
References
Huang MX, Mosher JC, Leahy RM (1999)
"A sensor-weighted overlapping-sphere head model and exhaustive head model comparison for MEG," Phys Med Biol, 44:423-440 | pdf
Additional documentation
Tutorial: BEM with OpenMEEG
Tutorial: Volume source estimation
External documentation: Electromagnetic neural source imaging
Forum: Sensor modeling: http://neuroimage.usc.edu/forums/showthread.php?1295
Forum: EEG reference: http://neuroimage.usc.edu/forums/showthread.php?1525#post6718
Forum: EEG and default anatomy: http://neuroimage.usc.edu/forums/showthread.php?1774
Forum: Mixed head models indices: http://neuroimage.usc.edu/forums/showthread.php?1878